Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
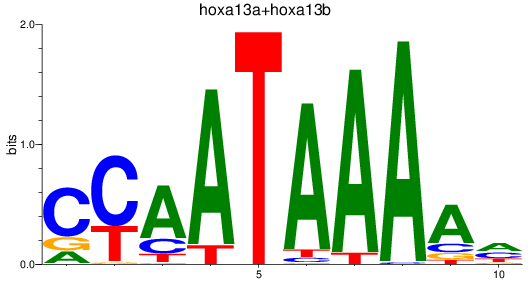
Results for hoxa13a+hoxa13b
Z-value: 0.64
Transcription factors associated with hoxa13a+hoxa13b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa13b
|
ENSDARG00000036254 | homeobox A13b |
|
hoxa13a
|
ENSDARG00000100312 | homeobox A13a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa13b | dr11_v1_chr16_+_20895904_20895904 | 0.59 | 9.6e-03 | Click! |
| hoxa13a | dr11_v1_chr19_+_19729506_19729506 | 0.48 | 4.5e-02 | Click! |
Activity profile of hoxa13a+hoxa13b motif
Sorted Z-values of hoxa13a+hoxa13b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_39606108 | 2.08 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr7_-_30367650 | 1.54 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr14_+_80685 | 1.48 |
ENSDART00000188443
|
stag3
|
stromal antigen 3 |
| chr6_+_13039951 | 1.30 |
ENSDART00000091700
|
catip
|
ciliogenesis associated TTC17 interacting protein |
| chr19_-_10881486 | 1.17 |
ENSDART00000168852
ENSDART00000160438 |
PSMD4 (1 of many)
psmd4a
|
proteasome 26S subunit, non-ATPase 4 proteasome 26S subunit, non-ATPase 4a |
| chr12_+_19356623 | 1.10 |
ENSDART00000078284
|
dmc1
|
DNA meiotic recombinase 1 |
| chr5_-_23200880 | 1.08 |
ENSDART00000051531
|
iqcd
|
IQ motif containing D |
| chr20_+_2139436 | 1.07 |
ENSDART00000155311
|
l3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr17_+_21295132 | 1.06 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr13_-_37619159 | 0.99 |
ENSDART00000186348
|
zgc:152791
|
zgc:152791 |
| chr21_+_22845317 | 0.99 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr25_+_34641536 | 0.99 |
ENSDART00000167033
|
CABZ01079011.1
|
|
| chr21_-_2299002 | 0.99 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr3_-_19200571 | 0.97 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr10_+_26612321 | 0.96 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr14_-_21123551 | 0.96 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr5_+_67971627 | 0.94 |
ENSDART00000144879
|
mtif3
|
mitochondrial translational initiation factor 3 |
| chr8_+_17933475 | 0.94 |
ENSDART00000100651
|
erich3
|
glutamate-rich 3 |
| chr23_+_19813677 | 0.93 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr20_-_50049099 | 0.92 |
ENSDART00000123634
|
NDUFB1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr16_+_4838808 | 0.92 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr6_+_7421898 | 0.91 |
ENSDART00000043946
|
ccdc65
|
coiled-coil domain containing 65 |
| chr3_-_7656059 | 0.90 |
ENSDART00000170917
|
junbb
|
JunB proto-oncogene, AP-1 transcription factor subunit b |
| chr12_+_2870671 | 0.89 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr21_-_37733287 | 0.88 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr25_-_10503043 | 0.88 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr22_-_547748 | 0.88 |
ENSDART00000037455
ENSDART00000140101 |
ccnd3
|
cyclin D3 |
| chr21_+_45366229 | 0.87 |
ENSDART00000029946
|
ube2b
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog) |
| chr4_-_5652030 | 0.86 |
ENSDART00000010903
|
rsph9
|
radial spoke head 9 homolog |
| chr5_-_36948586 | 0.84 |
ENSDART00000193606
|
h3f3c
|
H3 histone, family 3C |
| chr19_+_17385561 | 0.84 |
ENSDART00000141397
ENSDART00000143913 ENSDART00000133626 |
rpl15
|
ribosomal protein L15 |
| chr23_+_42810055 | 0.82 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr7_+_12371061 | 0.82 |
ENSDART00000053860
|
saxo2
|
stabilizer of axonemal microtubules 2 |
| chr2_-_53896300 | 0.81 |
ENSDART00000161221
|
capsla
|
calcyphosine-like a |
| chr24_+_40860320 | 0.80 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr21_-_25756119 | 0.80 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr6_-_2627488 | 0.79 |
ENSDART00000044089
ENSDART00000158333 ENSDART00000155109 |
hyi
|
hydroxypyruvate isomerase |
| chr3_+_16771797 | 0.79 |
ENSDART00000147418
|
lrrc3cb
|
leucine rich repeat containing 3Cb |
| chr13_+_37022601 | 0.78 |
ENSDART00000131800
ENSDART00000041300 |
esr2b
|
estrogen receptor 2b |
| chr19_+_20778011 | 0.77 |
ENSDART00000024208
|
nutf2l
|
nuclear transport factor 2, like |
| chr6_-_19683406 | 0.77 |
ENSDART00000158041
|
cfap52
|
cilia and flagella associated protein 52 |
| chr3_+_13600714 | 0.77 |
ENSDART00000162124
|
hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr11_-_1392468 | 0.77 |
ENSDART00000004423
|
iars
|
isoleucyl-tRNA synthetase |
| chr4_-_64703 | 0.76 |
ENSDART00000167851
|
CU856344.1
|
|
| chr15_-_20933574 | 0.75 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr17_-_15382704 | 0.74 |
ENSDART00000005313
|
zgc:85722
|
zgc:85722 |
| chr16_-_54471235 | 0.74 |
ENSDART00000061572
|
cyp11c1
|
cytochrome P450, family 11, subfamily C, polypeptide 1 |
| chr2_+_49569017 | 0.72 |
ENSDART00000109471
|
sema4e
|
semaphorin 4e |
| chr12_+_8074343 | 0.72 |
ENSDART00000124084
|
cabcoco1
|
ciliary associated calcium binding coiled-coil 1 |
| chr16_+_42830152 | 0.72 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr21_+_44557006 | 0.70 |
ENSDART00000170159
|
cmc4
|
C-x(9)-C motif containing 4 homolog (S. cerevisiae) |
| chr6_+_43015916 | 0.70 |
ENSDART00000064888
|
tcta
|
T cell leukemia translocation altered |
| chr16_+_41668474 | 0.69 |
ENSDART00000141293
ENSDART00000102721 |
ubxn11
|
UBX domain protein 11 |
| chr5_-_61970674 | 0.69 |
ENSDART00000143161
|
znf541
|
zinc finger protein 541 |
| chr16_+_42829735 | 0.69 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr10_-_34889053 | 0.68 |
ENSDART00000136966
|
ccdc169
|
coiled-coil domain containing 169 |
| chr8_+_37755099 | 0.68 |
ENSDART00000075708
ENSDART00000140966 |
got1l1
|
glutamic-oxaloacetic transaminase 1 like 1 |
| chr20_+_52546186 | 0.67 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr19_+_40177246 | 0.67 |
ENSDART00000049147
|
rbm48
|
RNA binding motif protein 48 |
| chr16_-_21785261 | 0.67 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr4_+_14926948 | 0.66 |
ENSDART00000019647
|
psmc2
|
proteasome 26S subunit, ATPase 2 |
| chr17_+_7513673 | 0.66 |
ENSDART00000156674
|
klhl10b.1
|
kelch-like family member 10b, tandem duplicate 1 |
| chr15_+_25683069 | 0.65 |
ENSDART00000148190
|
hic1
|
hypermethylated in cancer 1 |
| chr23_+_19182819 | 0.64 |
ENSDART00000131804
|
si:dkey-93l1.4
|
si:dkey-93l1.4 |
| chr3_-_37759969 | 0.64 |
ENSDART00000151105
ENSDART00000151208 |
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr22_-_910926 | 0.64 |
ENSDART00000180075
|
FP016205.1
|
|
| chr20_-_40487208 | 0.64 |
ENSDART00000075070
ENSDART00000142029 |
hsf2
|
heat shock transcription factor 2 |
| chr20_+_34717403 | 0.64 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr6_+_46431848 | 0.63 |
ENSDART00000181056
ENSDART00000144569 ENSDART00000064865 ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr14_-_15171435 | 0.63 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr13_+_17694845 | 0.62 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr7_+_59169081 | 0.62 |
ENSDART00000167980
|
ostc
|
oligosaccharyltransferase complex subunit |
| chr14_-_24391424 | 0.62 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr6_+_27514465 | 0.61 |
ENSDART00000128985
ENSDART00000079397 |
ryk
|
receptor-like tyrosine kinase |
| chr15_-_34567370 | 0.60 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr17_-_26610814 | 0.60 |
ENSDART00000133402
ENSDART00000016608 |
mrpl57
|
mitochondrial ribosomal protein L57 |
| chr20_-_22798794 | 0.60 |
ENSDART00000148084
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr11_-_12364122 | 0.59 |
ENSDART00000170129
|
zgc:174353
|
zgc:174353 |
| chr12_-_31457301 | 0.59 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr10_+_42678520 | 0.59 |
ENSDART00000182496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr12_+_26471712 | 0.58 |
ENSDART00000162115
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr2_-_5728843 | 0.58 |
ENSDART00000014020
|
sst2
|
somatostatin 2 |
| chr22_+_18188045 | 0.58 |
ENSDART00000140106
|
mef2b
|
myocyte enhancer factor 2b |
| chr24_+_24923166 | 0.58 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr5_-_1203455 | 0.57 |
ENSDART00000172177
|
surf4
|
surfeit gene 4 |
| chr6_-_43233700 | 0.57 |
ENSDART00000113706
|
trnt1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr19_+_7810028 | 0.57 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr3_-_55147731 | 0.57 |
ENSDART00000155871
ENSDART00000109016 ENSDART00000122904 |
hbae3
|
hemoglobin alpha embryonic-3 |
| chr16_-_45917322 | 0.57 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr20_+_33739059 | 0.57 |
ENSDART00000140361
|
ppil6
|
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr3_-_53091946 | 0.56 |
ENSDART00000187297
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr17_-_12407106 | 0.55 |
ENSDART00000183540
|
ankef1b
|
ankyrin repeat and EF-hand domain containing 1b |
| chr22_+_15624371 | 0.55 |
ENSDART00000124868
|
lpl
|
lipoprotein lipase |
| chr11_+_7580079 | 0.55 |
ENSDART00000091550
ENSDART00000193223 ENSDART00000193386 |
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr22_+_18187857 | 0.54 |
ENSDART00000166300
|
mef2b
|
myocyte enhancer factor 2b |
| chr10_+_2899108 | 0.54 |
ENSDART00000147031
|
erap1a
|
endoplasmic reticulum aminopeptidase 1a |
| chr7_-_26457208 | 0.54 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr16_-_27543024 | 0.54 |
ENSDART00000147737
ENSDART00000078283 |
tex10
|
testis expressed 10 |
| chr13_+_25380432 | 0.54 |
ENSDART00000038524
|
gsto1
|
glutathione S-transferase omega 1 |
| chr20_-_52338782 | 0.53 |
ENSDART00000109735
ENSDART00000132941 |
si:ch1073-287p18.1
|
si:ch1073-287p18.1 |
| chr17_-_10309766 | 0.53 |
ENSDART00000160994
|
ttc6
|
tetratricopeptide repeat domain 6 |
| chr2_+_14992879 | 0.53 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr19_+_636886 | 0.53 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr19_-_1948236 | 0.53 |
ENSDART00000163344
|
znrf2a
|
zinc and ring finger 2a |
| chr8_+_23809843 | 0.52 |
ENSDART00000099724
|
si:ch211-163l21.10
|
si:ch211-163l21.10 |
| chr5_+_58687541 | 0.52 |
ENSDART00000083015
ENSDART00000181902 |
ccdc84
|
coiled-coil domain containing 84 |
| chr2_-_24907741 | 0.52 |
ENSDART00000155013
|
si:dkey-149i17.11
|
si:dkey-149i17.11 |
| chr4_-_149334 | 0.52 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr13_+_30804367 | 0.51 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr17_-_17764801 | 0.51 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr21_-_43079161 | 0.51 |
ENSDART00000144151
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr13_+_24584401 | 0.50 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr24_+_37903448 | 0.50 |
ENSDART00000155476
|
ccdc78
|
coiled-coil domain containing 78 |
| chr22_+_24715282 | 0.50 |
ENSDART00000088027
ENSDART00000189054 ENSDART00000140430 |
ssx2ipb
|
synovial sarcoma, X breakpoint 2 interacting protein b |
| chr20_+_25586099 | 0.50 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr17_-_31695217 | 0.49 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr13_+_844150 | 0.49 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr7_+_23515966 | 0.49 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr13_+_45980163 | 0.49 |
ENSDART00000074547
ENSDART00000005195 |
bsdc1
|
BSD domain containing 1 |
| chr6_-_49526510 | 0.49 |
ENSDART00000128025
|
rps26l
|
ribosomal protein S26, like |
| chr6_-_17206028 | 0.48 |
ENSDART00000155756
|
pimr3
|
Pim proto-oncogene, serine/threonine kinase, related 3 |
| chr6_-_28943056 | 0.48 |
ENSDART00000065138
|
tbc1d23
|
TBC1 domain family, member 23 |
| chr19_-_32600823 | 0.48 |
ENSDART00000134149
ENSDART00000187858 |
zgc:91944
|
zgc:91944 |
| chr24_-_21674950 | 0.48 |
ENSDART00000123216
ENSDART00000046211 |
lnx2a
|
ligand of numb-protein X 2a |
| chr11_-_25539323 | 0.48 |
ENSDART00000155785
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr22_+_2937485 | 0.48 |
ENSDART00000082222
ENSDART00000135507 ENSDART00000143258 |
cep19
|
centrosomal protein 19 |
| chr16_-_52821023 | 0.48 |
ENSDART00000074718
|
spire1b
|
spire-type actin nucleation factor 1b |
| chr22_+_2409175 | 0.48 |
ENSDART00000141776
|
zgc:113220
|
zgc:113220 |
| chr14_+_15495088 | 0.47 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr5_+_23597144 | 0.47 |
ENSDART00000143929
ENSDART00000142420 ENSDART00000032909 |
kat5b
|
K(lysine) acetyltransferase 5b |
| chr9_+_18716485 | 0.47 |
ENSDART00000135125
|
serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr21_-_27195256 | 0.47 |
ENSDART00000133152
ENSDART00000065401 |
zgc:110782
|
zgc:110782 |
| chr21_+_4702773 | 0.47 |
ENSDART00000147404
|
si:dkey-102g19.3
|
si:dkey-102g19.3 |
| chr2_-_30135446 | 0.47 |
ENSDART00000141906
|
trpa1a
|
transient receptor potential cation channel, subfamily A, member 1a |
| chr22_+_1911269 | 0.46 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr21_+_22828500 | 0.46 |
ENSDART00000151109
|
si:rp71-1p14.7
|
si:rp71-1p14.7 |
| chr6_-_8295657 | 0.46 |
ENSDART00000185135
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr5_-_26181863 | 0.46 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr16_-_35585374 | 0.46 |
ENSDART00000180208
|
scmh1
|
Scm polycomb group protein homolog 1 |
| chr4_-_72573359 | 0.46 |
ENSDART00000130342
|
si:cabz01054396.2
|
si:cabz01054396.2 |
| chr22_-_2937503 | 0.45 |
ENSDART00000092991
ENSDART00000131110 |
pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr2_+_24352497 | 0.45 |
ENSDART00000134909
|
pimr68
|
Pim proto-oncogene, serine/threonine kinase, related 68 |
| chr25_-_16146851 | 0.45 |
ENSDART00000104043
|
dkk3b
|
dickkopf WNT signaling pathway inhibitor 3b |
| chr2_-_27975530 | 0.45 |
ENSDART00000020367
|
zgc:56556
|
zgc:56556 |
| chr1_+_50538839 | 0.45 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr25_-_8916913 | 0.45 |
ENSDART00000104629
ENSDART00000131748 |
furinb
|
furin (paired basic amino acid cleaving enzyme) b |
| chr24_+_21621654 | 0.45 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr10_+_42374770 | 0.45 |
ENSDART00000020000
|
COX5B
|
zgc:86599 |
| chr8_+_14381272 | 0.44 |
ENSDART00000057642
|
acbd6
|
acyl-CoA binding domain containing 6 |
| chr20_-_14012859 | 0.43 |
ENSDART00000152429
|
si:ch211-22i13.2
|
si:ch211-22i13.2 |
| chr16_+_30575901 | 0.43 |
ENSDART00000077231
|
mc2r
|
melanocortin 2 receptor |
| chr14_-_25042184 | 0.43 |
ENSDART00000131027
|
TMEM216
|
si:rp71-1d10.5 |
| chr21_-_29364166 | 0.43 |
ENSDART00000188602
|
BX537120.3
|
|
| chr11_+_25539698 | 0.43 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr2_-_16159203 | 0.42 |
ENSDART00000153480
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr8_+_53051701 | 0.42 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr9_-_42696408 | 0.42 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr8_-_44299247 | 0.41 |
ENSDART00000144497
|
piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr5_+_54555567 | 0.41 |
ENSDART00000171159
|
anapc2
|
anaphase promoting complex subunit 2 |
| chr9_-_48214216 | 0.41 |
ENSDART00000012938
|
phgdh
|
phosphoglycerate dehydrogenase |
| chr2_-_24962820 | 0.41 |
ENSDART00000182767
|
hltf
|
helicase-like transcription factor |
| chr3_-_60316118 | 0.41 |
ENSDART00000171458
|
si:ch211-214b16.2
|
si:ch211-214b16.2 |
| chr21_-_22827548 | 0.41 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr24_+_23202545 | 0.41 |
ENSDART00000143862
|
pex2
|
peroxisomal biogenesis factor 2 |
| chr21_+_11503212 | 0.41 |
ENSDART00000146701
|
si:dkey-184p9.7
|
si:dkey-184p9.7 |
| chr4_-_18850799 | 0.41 |
ENSDART00000151844
|
mcat
|
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr10_+_18952271 | 0.41 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr13_-_37620091 | 0.41 |
ENSDART00000135875
ENSDART00000193270 ENSDART00000018064 |
zgc:152791
|
zgc:152791 |
| chr5_+_43807003 | 0.41 |
ENSDART00000097625
|
zgc:158640
|
zgc:158640 |
| chr23_-_35694171 | 0.40 |
ENSDART00000077539
|
tuba1c
|
tubulin, alpha 1c |
| chr12_-_36521767 | 0.40 |
ENSDART00000110290
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr21_+_13150937 | 0.40 |
ENSDART00000102251
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr7_+_24494299 | 0.40 |
ENSDART00000087568
|
nelfa
|
negative elongation factor complex member A |
| chr10_-_5016997 | 0.40 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr16_+_21242491 | 0.40 |
ENSDART00000145886
|
osbpl3b
|
oxysterol binding protein-like 3b |
| chr17_-_49280308 | 0.40 |
ENSDART00000059658
ENSDART00000169207 |
tfb1m
|
transcription factor B1, mitochondrial |
| chr24_-_9960290 | 0.40 |
ENSDART00000143390
ENSDART00000092975 ENSDART00000184953 |
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr23_+_7379728 | 0.40 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr5_-_9073433 | 0.39 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr19_+_7835025 | 0.39 |
ENSDART00000026276
|
cks1b
|
CDC28 protein kinase regulatory subunit 1B |
| chr1_-_35924495 | 0.39 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr2_-_30182353 | 0.39 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr3_+_22059066 | 0.39 |
ENSDART00000155739
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr14_-_24761132 | 0.39 |
ENSDART00000146299
|
slit3
|
slit homolog 3 (Drosophila) |
| chr9_+_41080029 | 0.39 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr7_-_24364536 | 0.39 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr11_-_3343463 | 0.39 |
ENSDART00000066177
|
tuba2
|
tubulin, alpha 2 |
| chr2_+_26682913 | 0.38 |
ENSDART00000010683
ENSDART00000131411 ENSDART00000137933 |
impa1
|
inositol monophosphatase 1 |
| chr21_-_21781158 | 0.38 |
ENSDART00000113734
|
chrdl2
|
chordin-like 2 |
| chr2_+_19578446 | 0.38 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr7_+_49664174 | 0.38 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr22_-_35330532 | 0.38 |
ENSDART00000172654
|
FO818743.1
|
|
| chr1_-_13989643 | 0.38 |
ENSDART00000191046
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr24_-_12689571 | 0.38 |
ENSDART00000015517
|
pdcd6
|
programmed cell death 6 |
| chr22_+_14051245 | 0.38 |
ENSDART00000043711
ENSDART00000164259 |
aox6
|
aldehyde oxidase 6 |
| chr6_+_49551614 | 0.38 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr3_-_23596809 | 0.38 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr13_+_13668991 | 0.38 |
ENSDART00000148266
|
pimr48
|
Pim proto-oncogene, serine/threonine kinase, related 48 |
| chr19_+_9097013 | 0.37 |
ENSDART00000139781
|
si:ch211-81a5.5
|
si:ch211-81a5.5 |
| chr25_-_35360096 | 0.37 |
ENSDART00000154053
ENSDART00000171917 |
si:ch73-147o17.1
|
si:ch73-147o17.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa13a+hoxa13b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.3 | 0.9 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 1.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.2 | 1.0 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.2 | 0.7 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.2 | 0.9 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 0.6 | GO:2000193 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.2 | 0.5 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 0.7 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 0.7 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.2 | 0.5 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.4 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.7 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.7 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.5 | GO:0050955 | thermoception(GO:0050955) detection of temperature stimulus involved in thermoception(GO:0050960) detection of temperature stimulus involved in sensory perception(GO:0050961) |
| 0.1 | 1.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.4 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.1 | 0.4 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.9 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.1 | 0.8 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.4 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.7 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.3 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.1 | 1.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.5 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.1 | 0.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.4 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.3 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 0.4 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.3 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 0.3 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.4 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.5 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.2 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.2 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.3 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 1.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.5 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.2 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.6 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.3 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.4 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.6 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 1.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.2 | GO:1902746 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.6 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 0.5 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.2 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.9 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.2 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.1 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.3 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.2 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.1 | 0.2 | GO:0003250 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 1.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.2 | GO:0048855 | glossopharyngeal nerve development(GO:0021563) adenohypophysis morphogenesis(GO:0048855) |
| 0.0 | 0.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.3 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.5 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.4 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.3 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.3 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.8 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.0 | 0.1 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.7 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.3 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.5 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.4 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.4 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.2 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.2 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.1 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.2 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.0 | GO:0051793 | medium-chain fatty acid metabolic process(GO:0051791) medium-chain fatty acid catabolic process(GO:0051793) |
| 0.0 | 0.1 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.3 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.4 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0060155 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.6 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.2 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.1 | GO:0010934 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.3 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.6 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.2 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.9 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.2 | GO:0010890 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.0 | 0.2 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.7 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.8 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 5.3 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.0 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.0 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
| 0.0 | 0.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.6 | GO:0071696 | ectodermal placode development(GO:0071696) |
| 0.0 | 0.2 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.3 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.9 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.4 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.0 | GO:1901827 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.0 | 0.8 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0001534 | radial spoke(GO:0001534) |
| 0.2 | 0.8 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.2 | 0.5 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 0.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 1.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.8 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 1.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.9 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 1.0 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.0 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 0.7 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 0.5 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.2 | 0.5 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.2 | 0.7 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.2 | 0.7 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 0.5 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 1.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:0034246 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.1 | 0.8 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 0.4 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.3 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.3 | GO:0001729 | ceramide kinase activity(GO:0001729) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.9 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.2 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.0 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.2 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.1 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.3 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.2 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.1 | 0.3 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.6 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.6 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.3 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.4 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 1.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.9 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 1.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.7 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0051379 | epinephrine binding(GO:0051379) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0005111 | type 1 fibroblast growth factor receptor binding(GO:0005105) type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 2.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.0 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.0 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.0 | 0.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 2.8 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 1.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |