Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for hoxa11b
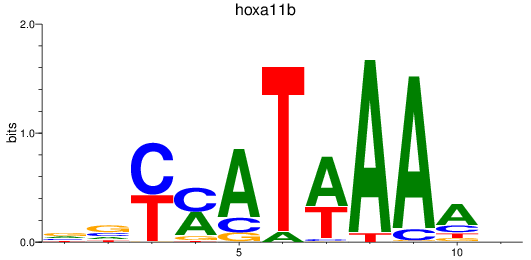
Z-value: 1.17
Transcription factors associated with hoxa11b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa11b
|
ENSDARG00000007009 | homeobox A11b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa11b | dr11_v1_chr16_+_20904754_20904754 | -0.71 | 8.6e-04 | Click! |
Activity profile of hoxa11b motif
Sorted Z-values of hoxa11b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_28208973 | 3.94 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr7_+_1473929 | 3.89 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr16_+_29509133 | 3.13 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr23_+_2666944 | 3.00 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr3_-_26184018 | 2.94 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr3_-_26183699 | 2.85 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr2_+_16597011 | 2.80 |
ENSDART00000160641
ENSDART00000125413 |
xrn1
|
5'-3' exoribonuclease 1 |
| chr1_-_6028876 | 2.69 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr8_+_12951155 | 2.67 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr14_+_32918172 | 2.64 |
ENSDART00000182867
|
lnx2b
|
ligand of numb-protein X 2b |
| chr24_-_10014512 | 2.63 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr8_+_12118097 | 2.56 |
ENSDART00000081819
|
endog
|
endonuclease G |
| chr21_-_19918286 | 2.53 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr2_-_32738535 | 2.50 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr12_-_4249000 | 2.25 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr5_+_44805028 | 2.24 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr6_+_4528631 | 2.24 |
ENSDART00000122042
|
RNF219
|
ring finger protein 219 |
| chr14_+_34490445 | 2.20 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr1_+_494297 | 2.20 |
ENSDART00000108579
ENSDART00000146732 |
blzf1
|
basic leucine zipper nuclear factor 1 |
| chr5_+_44805269 | 2.20 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr19_-_47571456 | 2.16 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr20_-_51831657 | 2.05 |
ENSDART00000165076
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr20_-_34028967 | 2.00 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr20_-_51831816 | 1.97 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr13_+_33462232 | 1.93 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr5_+_3891485 | 1.92 |
ENSDART00000129329
ENSDART00000091711 |
rpain
|
RPA interacting protein |
| chr12_-_25887864 | 1.92 |
ENSDART00000152983
|
si:dkey-193p11.2
|
si:dkey-193p11.2 |
| chr10_-_10969596 | 1.89 |
ENSDART00000092011
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr6_-_9282080 | 1.88 |
ENSDART00000159506
|
ccdc14
|
coiled-coil domain containing 14 |
| chr21_-_22357545 | 1.84 |
ENSDART00000134320
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr8_+_45334255 | 1.82 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr20_-_49889111 | 1.81 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr6_-_42388608 | 1.81 |
ENSDART00000049425
|
sec61a1l
|
Sec61 translocon alpha 1 subunit, like |
| chr8_+_21229718 | 1.78 |
ENSDART00000100222
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr19_+_9111550 | 1.78 |
ENSDART00000088336
|
setdb1a
|
SET domain, bifurcated 1a |
| chr17_+_14965570 | 1.77 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr19_-_31042570 | 1.73 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr22_-_11661724 | 1.72 |
ENSDART00000025198
|
mettl21a
|
methyltransferase like 21A |
| chr22_+_10713713 | 1.71 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr3_+_46763745 | 1.71 |
ENSDART00000185437
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr12_+_22674030 | 1.68 |
ENSDART00000153254
ENSDART00000152930 |
cdca9
|
cell division cycle associated 9 |
| chr16_+_23487051 | 1.67 |
ENSDART00000145496
|
icn2
|
ictacalcin 2 |
| chr10_-_10969444 | 1.67 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr14_-_33481428 | 1.65 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr19_-_10915898 | 1.65 |
ENSDART00000163179
|
pip5k1aa
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, a |
| chr12_-_10512911 | 1.64 |
ENSDART00000124562
ENSDART00000106163 |
zgc:152977
|
zgc:152977 |
| chr11_-_6452444 | 1.64 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr22_-_10541372 | 1.62 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr3_-_3366590 | 1.62 |
ENSDART00000109428
ENSDART00000175329 |
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr12_+_16087077 | 1.61 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr25_+_7532627 | 1.59 |
ENSDART00000187660
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr2_-_44777592 | 1.57 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr4_+_20486041 | 1.56 |
ENSDART00000017572
|
ints13
|
integrator complex subunit 13 |
| chr14_+_15155684 | 1.55 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr13_-_2981472 | 1.55 |
ENSDART00000184820
|
CABZ01087623.1
|
|
| chr20_-_29499363 | 1.54 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr5_-_1274794 | 1.53 |
ENSDART00000167337
ENSDART00000125247 |
ciz1a
|
cdkn1a interacting zinc finger protein 1a |
| chr4_-_20108833 | 1.50 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr19_-_47571797 | 1.50 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr17_-_4245902 | 1.49 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr13_+_36585399 | 1.46 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr7_-_69429561 | 1.46 |
ENSDART00000127351
|
atxn1l
|
ataxin 1-like |
| chr3_+_39579393 | 1.46 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr16_-_41717063 | 1.45 |
ENSDART00000084610
|
cep85
|
centrosomal protein 85 |
| chr13_-_4018888 | 1.44 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr15_-_16946124 | 1.44 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr12_-_17152139 | 1.43 |
ENSDART00000152478
|
stambpl1
|
STAM binding protein-like 1 |
| chr19_-_27550768 | 1.42 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr15_+_29024895 | 1.41 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr18_-_3520358 | 1.41 |
ENSDART00000181412
|
capn5a
|
calpain 5a |
| chr13_-_24717618 | 1.40 |
ENSDART00000172156
|
erlin1
|
ER lipid raft associated 1 |
| chr17_+_24318753 | 1.40 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr12_+_19408373 | 1.39 |
ENSDART00000114248
|
snx29
|
sorting nexin 29 |
| chr7_+_71955486 | 1.38 |
ENSDART00000189349
|
CABZ01071171.1
|
Danio rerio low density lipoprotein receptor-related protein 4 (lrp4), mRNA. |
| chr12_-_20616160 | 1.38 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr5_-_54712159 | 1.38 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr3_+_24062748 | 1.37 |
ENSDART00000188574
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr11_-_25257045 | 1.37 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr5_-_36597612 | 1.36 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr13_+_39277178 | 1.36 |
ENSDART00000113259
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr1_-_55044256 | 1.36 |
ENSDART00000165505
ENSDART00000167536 ENSDART00000170001 |
vps54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr2_-_45663945 | 1.35 |
ENSDART00000075080
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr2_+_3823813 | 1.35 |
ENSDART00000103596
ENSDART00000161880 ENSDART00000185408 |
npc1
|
Niemann-Pick disease, type C1 |
| chr7_-_24112484 | 1.35 |
ENSDART00000111923
|
ajuba
|
ajuba LIM protein |
| chr17_-_30521043 | 1.33 |
ENSDART00000087111
|
itsn2b
|
intersectin 2b |
| chr15_-_1843831 | 1.33 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr10_+_15967341 | 1.32 |
ENSDART00000115130
ENSDART00000189687 |
apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr5_+_44846280 | 1.31 |
ENSDART00000084370
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr12_-_33817114 | 1.31 |
ENSDART00000161265
|
twnk
|
twinkle mtDNA helicase |
| chr1_+_44173245 | 1.30 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr16_+_40340222 | 1.30 |
ENSDART00000190631
|
mettl6
|
methyltransferase like 6 |
| chr5_-_54554583 | 1.30 |
ENSDART00000158865
ENSDART00000158069 |
ssna1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr12_-_44759246 | 1.30 |
ENSDART00000162888
|
dock1
|
dedicator of cytokinesis 1 |
| chr7_-_51773166 | 1.29 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr5_-_15283509 | 1.29 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr9_+_2452672 | 1.29 |
ENSDART00000193993
|
chn1
|
chimerin 1 |
| chr2_+_22409249 | 1.29 |
ENSDART00000182915
|
zgc:56628
|
zgc:56628 |
| chr11_+_24313931 | 1.28 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr21_-_32781612 | 1.28 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr16_+_38940758 | 1.28 |
ENSDART00000102482
ENSDART00000136215 |
eny2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr11_+_11303458 | 1.26 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr5_-_65662996 | 1.26 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr8_-_13722682 | 1.26 |
ENSDART00000142052
|
si:dkey-258f14.2
|
si:dkey-258f14.2 |
| chr17_+_21887823 | 1.26 |
ENSDART00000131929
ENSDART00000165192 |
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr10_+_15967643 | 1.26 |
ENSDART00000136709
|
apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr7_+_32695954 | 1.25 |
ENSDART00000184425
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr5_-_48664522 | 1.25 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr9_+_32178374 | 1.24 |
ENSDART00000078576
|
coq10b
|
coenzyme Q10B |
| chr16_-_25368048 | 1.24 |
ENSDART00000132445
|
rbfa
|
ribosome binding factor A |
| chr25_+_7532811 | 1.24 |
ENSDART00000161593
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr6_-_41138854 | 1.24 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr9_-_41040492 | 1.23 |
ENSDART00000163164
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr16_-_28658341 | 1.23 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr13_+_47710434 | 1.23 |
ENSDART00000188724
|
TMEM87B
|
transmembrane protein 87B |
| chr1_+_2301961 | 1.23 |
ENSDART00000108919
ENSDART00000143361 ENSDART00000142944 |
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr3_-_32337653 | 1.22 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr18_+_26428829 | 1.22 |
ENSDART00000190779
ENSDART00000193226 ENSDART00000110746 |
blm
|
Bloom syndrome, RecQ helicase-like |
| chr1_+_2321384 | 1.22 |
ENSDART00000157662
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr18_-_12295092 | 1.22 |
ENSDART00000033248
|
fam107b
|
family with sequence similarity 107, member B |
| chr21_+_13327527 | 1.22 |
ENSDART00000114294
|
snrpd3l
|
small nuclear ribonucleoprotein D3 polypeptide, like |
| chr16_+_39146696 | 1.21 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr20_+_29565906 | 1.21 |
ENSDART00000062383
|
ywhaqa
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide a |
| chr8_+_23355484 | 1.21 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr17_+_17764979 | 1.20 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr15_+_1534644 | 1.20 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr22_-_10158038 | 1.20 |
ENSDART00000047444
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr20_+_14789305 | 1.20 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr10_+_15454745 | 1.19 |
ENSDART00000129441
ENSDART00000123935 ENSDART00000163446 ENSDART00000087680 ENSDART00000193752 |
erbin
|
erbb2 interacting protein |
| chr20_-_25643667 | 1.19 |
ENSDART00000137457
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr8_-_30395888 | 1.18 |
ENSDART00000144637
|
dock8
|
dedicator of cytokinesis 8 |
| chr24_-_26622423 | 1.18 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr22_-_881725 | 1.17 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr10_+_5234327 | 1.17 |
ENSDART00000133927
ENSDART00000063120 |
sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr24_-_36238054 | 1.17 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr25_-_2723902 | 1.17 |
ENSDART00000143721
ENSDART00000175224 |
adpgk
|
ADP-dependent glucokinase |
| chr14_+_45028062 | 1.17 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr24_+_29912509 | 1.16 |
ENSDART00000168422
|
frrs1b
|
ferric-chelate reductase 1b |
| chr8_-_39822917 | 1.16 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr3_+_46764022 | 1.15 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr18_+_15876385 | 1.15 |
ENSDART00000142527
|
eea1
|
early endosome antigen 1 |
| chr12_+_30788912 | 1.15 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr16_+_7380463 | 1.15 |
ENSDART00000029727
ENSDART00000149086 |
atg5
|
ATG5 autophagy related 5 homolog (S. cerevisiae) |
| chr23_+_43718115 | 1.14 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr20_-_34670236 | 1.14 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr15_-_26931541 | 1.14 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr21_+_3166519 | 1.14 |
ENSDART00000160585
|
hnrnpabb
|
heterogeneous nuclear ribonucleoprotein A/Bb |
| chr5_-_48070779 | 1.13 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr21_-_22122312 | 1.13 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr14_+_16036139 | 1.12 |
ENSDART00000190733
|
prelid1a
|
PRELI domain containing 1a |
| chr15_+_25439106 | 1.12 |
ENSDART00000156252
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr12_-_17686404 | 1.12 |
ENSDART00000079065
|
ccz1
|
CCZ1 homolog, vacuolar protein trafficking and biogenesis associated |
| chr8_+_50190742 | 1.12 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr22_+_2751887 | 1.11 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr15_-_23721618 | 1.11 |
ENSDART00000109318
|
zc3h4
|
zinc finger CCCH-type containing 4 |
| chr2_-_21170517 | 1.10 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr16_-_10223741 | 1.10 |
ENSDART00000188099
|
si:rp71-15i12.1
|
si:rp71-15i12.1 |
| chr5_-_62940851 | 1.10 |
ENSDART00000137052
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr9_-_30259295 | 1.09 |
ENSDART00000139106
|
si:dkey-100n23.5
|
si:dkey-100n23.5 |
| chr24_-_33801668 | 1.09 |
ENSDART00000079202
|
abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr24_+_36840652 | 1.09 |
ENSDART00000088168
|
CABZ01055347.1
|
|
| chr8_+_9699111 | 1.08 |
ENSDART00000111853
|
gripap1
|
GRIP1 associated protein 1 |
| chr19_-_30447611 | 1.08 |
ENSDART00000073705
ENSDART00000048977 ENSDART00000191237 |
abcf1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr21_+_3166988 | 1.08 |
ENSDART00000168163
|
hnrnpabb
|
heterogeneous nuclear ribonucleoprotein A/Bb |
| chr9_-_34269066 | 1.08 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr16_-_42965192 | 1.08 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr20_-_48700370 | 1.08 |
ENSDART00000185195
ENSDART00000184898 |
pax1b
|
paired box 1b |
| chr10_+_8197827 | 1.07 |
ENSDART00000026244
|
mtrex
|
Mtr4 exosome RNA helicase |
| chr8_-_31416403 | 1.07 |
ENSDART00000098912
|
znf131
|
zinc finger protein 131 |
| chr13_-_23067381 | 1.07 |
ENSDART00000078140
|
vps26a
|
vacuolar protein sorting 26 homolog A (S. pombe) |
| chr6_-_33685325 | 1.07 |
ENSDART00000181883
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr5_-_29531948 | 1.07 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr16_+_40340523 | 1.06 |
ENSDART00000102571
|
mettl6
|
methyltransferase like 6 |
| chr18_+_18000695 | 1.06 |
ENSDART00000146898
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr3_-_32873641 | 1.05 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr25_-_25142387 | 1.05 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr5_+_67835595 | 1.05 |
ENSDART00000159386
|
si:ch211-271b14.1
|
si:ch211-271b14.1 |
| chr19_-_432083 | 1.04 |
ENSDART00000165371
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr5_-_11809710 | 1.04 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr3_+_17933132 | 1.04 |
ENSDART00000104299
ENSDART00000162144 ENSDART00000162242 ENSDART00000166289 ENSDART00000171101 ENSDART00000164853 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr22_-_31517300 | 1.03 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr14_+_14841685 | 1.03 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr8_+_13364950 | 1.02 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr3_-_43650189 | 1.02 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr8_+_35964482 | 1.02 |
ENSDART00000129357
ENSDART00000154953 |
glt1d1
|
glycosyltransferase 1 domain containing 1 |
| chr23_+_39963599 | 1.02 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr3_-_36364903 | 1.02 |
ENSDART00000028883
|
gna13b
|
guanine nucleotide binding protein (G protein), alpha 13b |
| chr10_+_15608326 | 1.02 |
ENSDART00000188770
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr16_+_13883872 | 1.02 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr16_-_41646164 | 1.02 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr1_+_6225493 | 1.01 |
ENSDART00000145378
|
fastkd2
|
FAST kinase domains 2 |
| chr12_-_13650344 | 1.01 |
ENSDART00000124364
ENSDART00000124638 ENSDART00000171929 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr8_-_28274552 | 1.01 |
ENSDART00000131580
|
rap1aa
|
RAP1A, member of RAS oncogene family a |
| chr17_-_10498347 | 1.01 |
ENSDART00000171324
|
mipol1
|
mirror-image polydactyly 1 |
| chr19_+_40069524 | 1.00 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr5_+_9037650 | 1.00 |
ENSDART00000158226
|
si:ch211-155m12.1
|
si:ch211-155m12.1 |
| chr13_+_22675802 | 1.00 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr4_-_1776352 | 0.99 |
ENSDART00000123089
|
dnajc2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr5_-_39805874 | 0.99 |
ENSDART00000176202
ENSDART00000191683 |
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr3_+_26244353 | 0.99 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr16_+_35916371 | 0.99 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa11b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.7 | 2.2 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.6 | 1.7 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.5 | 3.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.5 | 5.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.5 | 5.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.4 | 1.3 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.4 | 1.2 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.4 | 1.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 1.2 | GO:0097065 | anterior head development(GO:0097065) |
| 0.4 | 1.9 | GO:2000301 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.4 | 2.6 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.3 | 3.4 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.3 | 1.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.3 | 1.3 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.3 | 1.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.3 | 1.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.3 | 1.5 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.3 | 2.0 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.3 | 0.8 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.3 | 1.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.3 | 1.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.3 | 1.8 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 1.3 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 1.0 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.3 | 1.0 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.3 | 0.8 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 1.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.2 | 1.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 0.7 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.2 | 0.9 | GO:1904590 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.2 | 0.7 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 2.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 4.0 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.2 | 1.1 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.2 | 3.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.2 | 1.5 | GO:0015809 | arginine transport(GO:0015809) |
| 0.2 | 1.9 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 1.0 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.2 | 1.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 2.4 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 3.0 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.2 | 0.7 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 1.8 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.2 | 0.5 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.2 | 0.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 0.7 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.2 | 1.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 0.5 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.2 | 0.5 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.2 | 1.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.2 | 1.8 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.2 | 2.4 | GO:1900151 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.2 | 0.6 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 0.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 3.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 1.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 0.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 0.9 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 1.0 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) histone mRNA metabolic process(GO:0008334) |
| 0.1 | 1.8 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.9 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.6 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 0.5 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.7 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.3 | GO:0070228 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.1 | 1.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.5 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 2.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.6 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 1.0 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.8 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.6 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 0.7 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.3 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.6 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.1 | 0.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 1.4 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 1.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 1.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.5 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 0.8 | GO:0035912 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 1.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.9 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.3 | GO:0002631 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.5 | GO:0097094 | regulation of osteoblast proliferation(GO:0033688) craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.8 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.4 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 1.0 | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 0.1 | 0.3 | GO:0030238 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.1 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.4 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 1.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.7 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.1 | 0.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 2.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 1.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 2.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 1.0 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.5 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 1.9 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.1 | 0.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.4 | GO:0009988 | cell-cell recognition(GO:0009988) |
| 0.1 | 1.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 1.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.4 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 1.3 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.1 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.7 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 1.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.1 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.4 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.1 | 1.2 | GO:1901661 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 2.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 1.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 1.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 1.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.5 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.9 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.1 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 0.5 | GO:0071174 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.1 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 1.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.2 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.1 | 0.3 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 0.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 0.2 | GO:1900180 | regulation of protein localization to nucleus(GO:1900180) |
| 0.1 | 0.9 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 2.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.2 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 1.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.1 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.3 | GO:1900027 | regulation of ruffle assembly(GO:1900027) positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 1.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.8 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.8 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) |
| 0.0 | 0.8 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 1.3 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.7 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.3 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.5 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.0 | 0.8 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.2 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.2 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 1.3 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 1.1 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.9 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 1.8 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 1.2 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0070169 | positive regulation of bone mineralization(GO:0030501) positive regulation of ossification(GO:0045778) positive regulation of biomineral tissue development(GO:0070169) |
| 0.0 | 0.8 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 2.3 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.5 | GO:0051383 | kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.0 | 1.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 5.3 | GO:0007034 | vacuolar transport(GO:0007034) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 1.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.6 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 1.5 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.6 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.3 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 2.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.8 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.0 | 0.0 | GO:0032640 | tumor necrosis factor production(GO:0032640) positive regulation of interleukin-6 production(GO:0032755) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 0.5 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.4 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.3 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.8 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.7 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.2 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.7 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 4.8 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 1.1 | GO:0018210 | peptidyl-threonine modification(GO:0018210) |
| 0.0 | 0.2 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.1 | GO:2000725 | regulation of cardiac muscle tissue development(GO:0055024) regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.1 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of hormone metabolic process(GO:0032350) histone H3-K4 demethylation(GO:0034720) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.5 | GO:0034111 | negative regulation of homotypic cell-cell adhesion(GO:0034111) negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 1.0 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.3 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.2 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.1 | GO:0019859 | thymine catabolic process(GO:0006210) uracil catabolic process(GO:0006212) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.0 | 0.9 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.4 | GO:0032231 | regulation of actin filament bundle assembly(GO:0032231) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.2 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 1.9 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.3 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.4 | GO:0031647 | regulation of protein stability(GO:0031647) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.7 | 2.9 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.6 | 1.7 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.5 | 1.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.4 | 1.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.3 | 1.8 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.3 | 1.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 1.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 2.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 1.3 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.3 | 1.8 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 0.9 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.2 | 1.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 0.6 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 1.8 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 0.9 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 1.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.2 | 1.0 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.2 | 1.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 0.5 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 3.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 1.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 2.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 3.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.4 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.6 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.9 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.7 | GO:0001650 | fibrillar center(GO:0001650) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 2.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.6 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.3 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.6 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.4 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 0.8 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 4.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.6 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 2.3 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.6 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 2.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.2 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 1.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 10.0 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.1 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 2.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 2.2 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.0 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 3.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.3 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 6.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.7 | 5.2 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.7 | 2.8 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 2.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.4 | 1.6 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.4 | 1.2 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.4 | 1.2 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.4 | 1.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 1.0 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.3 | 1.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 1.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.3 | 1.2 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.3 | 0.9 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.3 | 2.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.3 | 1.3 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 1.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 1.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 1.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.2 | 1.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 0.6 | GO:0000996 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.2 | 1.4 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.2 | 0.8 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.2 | 0.6 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.2 | 1.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.2 | 0.7 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.2 | 1.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 0.9 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 0.7 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 0.5 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.2 | 1.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 0.7 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.2 | 1.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 2.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 1.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.5 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 1.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 2.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 1.8 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 2.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.4 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.1 | 0.7 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.6 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 1.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 1.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.7 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 1.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.7 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.6 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 2.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.4 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 1.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 1.0 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 3.6 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 2.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.5 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.6 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.4 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.3 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.4 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.2 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.1 | 0.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 7.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 2.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.8 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 1.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.7 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.0 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 1.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 1.0 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.6 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.8 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.4 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 1.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.6 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.6 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 2.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.9 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.9 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.1 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 1.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.1 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.0 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.0 | GO:0008168 | methyltransferase activity(GO:0008168) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 4.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 4.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.8 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 3.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.2 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 2.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 5.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 3.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 1.8 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.2 | 1.1 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.2 | 2.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 1.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 0.8 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 2.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 2.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.9 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.1 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |