Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
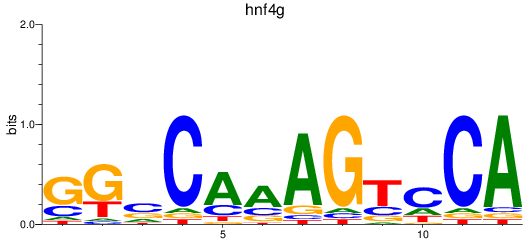
Results for hnf4g
Z-value: 1.29
Transcription factors associated with hnf4g
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hnf4g
|
ENSDARG00000071565 | hepatocyte nuclear factor 4, gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hnf4g | dr11_v1_chr24_-_23671709_23671709 | -0.79 | 9.8e-05 | Click! |
Activity profile of hnf4g motif
Sorted Z-values of hnf4g motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_2590222 | 7.22 |
ENSDART00000185711
|
CR759892.1
|
|
| chr17_-_2573021 | 6.13 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr17_-_2578026 | 6.02 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr17_-_2595736 | 5.62 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr17_-_2584423 | 5.57 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr4_-_77114795 | 5.50 |
ENSDART00000144849
|
CU467646.2
|
|
| chr10_+_19569052 | 5.39 |
ENSDART00000058425
|
CABZ01059627.1
|
|
| chr9_+_8380728 | 5.03 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr22_-_24992532 | 2.90 |
ENSDART00000102751
|
si:dkey-179j5.5
|
si:dkey-179j5.5 |
| chr16_+_26846495 | 2.81 |
ENSDART00000078124
|
trim35-29
|
tripartite motif containing 35-29 |
| chr23_+_2740741 | 2.39 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr7_-_24520866 | 2.33 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr13_-_35908275 | 1.71 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr5_-_30080332 | 1.69 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr13_-_35907768 | 1.65 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr15_-_30857350 | 1.54 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr7_-_26518086 | 1.53 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr14_-_16810401 | 1.50 |
ENSDART00000158396
ENSDART00000170758 |
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr1_-_12064715 | 1.39 |
ENSDART00000143628
ENSDART00000103406 |
pla2g12a
|
phospholipase A2, group XIIA |
| chr8_-_49207319 | 1.38 |
ENSDART00000022870
|
fam110a
|
family with sequence similarity 110, member A |
| chr1_-_9485939 | 1.36 |
ENSDART00000157814
|
micall2b
|
mical-like 2b |
| chr5_-_69180587 | 1.35 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr17_-_6076266 | 1.31 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr15_+_29728377 | 1.25 |
ENSDART00000099958
|
zgc:153372
|
zgc:153372 |
| chr8_-_1264893 | 1.22 |
ENSDART00000190371
|
cdc14b
|
cell division cycle 14B |
| chr5_-_69180227 | 1.21 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr17_-_6076084 | 1.20 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr7_-_59514547 | 1.19 |
ENSDART00000168457
|
slx1b
|
SLX1 homolog B, structure-specific endonuclease subunit |
| chr7_-_20611039 | 1.15 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr7_+_24520518 | 1.12 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr11_-_18017287 | 1.06 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr6_-_31682135 | 1.02 |
ENSDART00000153988
|
cachd1
|
cache domain containing 1 |
| chr11_-_5563498 | 1.02 |
ENSDART00000160835
|
pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr2_-_42173834 | 0.97 |
ENSDART00000098357
ENSDART00000144707 |
slc39a6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr3_-_36440705 | 0.93 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr11_-_27827442 | 0.90 |
ENSDART00000121847
ENSDART00000132018 ENSDART00000145744 ENSDART00000134677 ENSDART00000130800 |
cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1 |
| chr5_-_36597612 | 0.90 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr7_+_29115890 | 0.83 |
ENSDART00000052345
|
tradd
|
tnfrsf1a-associated via death domain |
| chr13_+_32807419 | 0.79 |
ENSDART00000085252
|
pqlc3
|
PQ loop repeat containing 3 |
| chr19_-_1002959 | 0.75 |
ENSDART00000168138
|
ehmt2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr25_-_24240797 | 0.74 |
ENSDART00000132790
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr12_-_10567188 | 0.74 |
ENSDART00000144283
|
myof
|
myoferlin |
| chr24_+_11083146 | 0.70 |
ENSDART00000009473
|
zfand1
|
zinc finger, AN1-type domain 1 |
| chr3_-_27868183 | 0.70 |
ENSDART00000185812
|
abat
|
4-aminobutyrate aminotransferase |
| chr20_+_34151670 | 0.69 |
ENSDART00000152870
ENSDART00000010329 ENSDART00000145852 |
arpc5b
|
actin related protein 2/3 complex, subunit 5B |
| chr22_-_31517300 | 0.66 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr4_+_11172964 | 0.66 |
ENSDART00000193598
ENSDART00000138661 |
tspan11
|
tetraspanin 11 |
| chr22_-_24738188 | 0.66 |
ENSDART00000050238
|
vtg1
|
vitellogenin 1 |
| chr7_+_24496894 | 0.64 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr6_-_46474483 | 0.62 |
ENSDART00000155761
|
rdh20
|
retinol dehydrogenase 20 |
| chr24_-_36175365 | 0.57 |
ENSDART00000065338
|
pak1ip1
|
PAK1 interacting protein 1 |
| chr21_-_43474012 | 0.53 |
ENSDART00000065104
|
tmem185
|
transmembrane protein 185 |
| chr7_-_29115772 | 0.51 |
ENSDART00000076386
|
fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr1_-_57172294 | 0.48 |
ENSDART00000063774
|
rac1l
|
Rac family small GTPase 1, like |
| chr15_+_34069746 | 0.47 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr8_-_20838342 | 0.46 |
ENSDART00000141345
|
si:ch211-133l5.7
|
si:ch211-133l5.7 |
| chr15_-_25153352 | 0.44 |
ENSDART00000078095
ENSDART00000122184 |
vps53
|
vacuolar protein sorting 53 homolog (S. cerevisiae) |
| chr11_+_12860260 | 0.44 |
ENSDART00000190410
|
zgpat
|
zinc finger, CCCH-type with G patch domain |
| chr11_+_19068442 | 0.43 |
ENSDART00000171766
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr6_-_10788065 | 0.41 |
ENSDART00000190968
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr16_+_48460873 | 0.40 |
ENSDART00000159902
|
ext1a
|
exostosin glycosyltransferase 1a |
| chr21_-_5879897 | 0.38 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr24_+_7828097 | 0.36 |
ENSDART00000134975
|
zgc:101569
|
zgc:101569 |
| chr15_+_8192715 | 0.33 |
ENSDART00000156442
ENSDART00000077660 |
mipep
|
mitochondrial intermediate peptidase |
| chr4_-_170120 | 0.32 |
ENSDART00000171333
|
eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr10_+_45128375 | 0.32 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr12_+_33916939 | 0.31 |
ENSDART00000149292
|
trim8b
|
tripartite motif containing 8b |
| chr10_-_24753715 | 0.28 |
ENSDART00000192401
|
ilk
|
integrin-linked kinase |
| chr11_+_22134540 | 0.26 |
ENSDART00000190502
|
MDFI
|
si:dkey-91m3.1 |
| chr6_-_12296170 | 0.26 |
ENSDART00000155685
|
pkp4
|
plakophilin 4 |
| chr20_+_29436601 | 0.26 |
ENSDART00000136804
|
fmn1
|
formin 1 |
| chr21_-_27272657 | 0.24 |
ENSDART00000040754
ENSDART00000175009 |
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr20_-_46128590 | 0.23 |
ENSDART00000123744
|
taar1b
|
trace amine associated receptor 1b |
| chr15_+_3825117 | 0.22 |
ENSDART00000183315
|
CABZ01061591.1
|
|
| chr21_-_11054876 | 0.22 |
ENSDART00000146576
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr15_-_2632891 | 0.22 |
ENSDART00000081840
|
cldnj
|
claudin j |
| chr19_+_10536093 | 0.20 |
ENSDART00000138706
|
si:dkey-211g8.1
|
si:dkey-211g8.1 |
| chr2_-_10877765 | 0.19 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr24_+_25467465 | 0.19 |
ENSDART00000189933
|
smpx
|
small muscle protein, X-linked |
| chr22_+_2844865 | 0.18 |
ENSDART00000139123
|
si:dkey-20i20.4
|
si:dkey-20i20.4 |
| chr15_-_41700406 | 0.18 |
ENSDART00000181460
|
si:ch211-276c2.4
|
si:ch211-276c2.4 |
| chr19_-_25519310 | 0.17 |
ENSDART00000089882
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr22_+_1300587 | 0.17 |
ENSDART00000124161
|
si:ch73-138e16.5
|
si:ch73-138e16.5 |
| chr20_-_46694573 | 0.17 |
ENSDART00000182557
|
AL929435.2
|
|
| chr18_-_9016161 | 0.17 |
ENSDART00000189888
|
grm3
|
glutamate receptor, metabotropic 3 |
| chr7_+_17229282 | 0.16 |
ENSDART00000097982
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr5_-_62317496 | 0.16 |
ENSDART00000180089
|
zgc:85789
|
zgc:85789 |
| chr23_-_43595956 | 0.16 |
ENSDART00000162186
|
itchb
|
itchy E3 ubiquitin protein ligase b |
| chr15_+_864761 | 0.14 |
ENSDART00000174813
ENSDART00000192053 |
si:dkey-7i4.12
si:dkey-7i4.10
|
si:dkey-7i4.12 si:dkey-7i4.10 |
| chr15_-_16012963 | 0.14 |
ENSDART00000144138
|
hnf1ba
|
HNF1 homeobox Ba |
| chr22_-_24818066 | 0.13 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr6_-_56103718 | 0.13 |
ENSDART00000191002
|
gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr9_-_1986014 | 0.12 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr25_+_22216723 | 0.12 |
ENSDART00000051291
|
hexa
|
hexosaminidase A (alpha polypeptide) |
| chr4_+_7888047 | 0.12 |
ENSDART00000104676
|
camk1da
|
calcium/calmodulin-dependent protein kinase 1Da |
| chr10_-_41400049 | 0.11 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr7_-_52417060 | 0.11 |
ENSDART00000148579
|
myzap
|
myocardial zonula adherens protein |
| chr4_+_53145525 | 0.11 |
ENSDART00000166960
|
si:dkey-8o9.2
|
si:dkey-8o9.2 |
| chr20_-_49657134 | 0.11 |
ENSDART00000151248
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr9_+_1138802 | 0.11 |
ENSDART00000191130
ENSDART00000187308 |
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr16_+_11623956 | 0.10 |
ENSDART00000137788
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr5_-_50640171 | 0.10 |
ENSDART00000183353
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr8_+_23390844 | 0.10 |
ENSDART00000184556
|
si:dkey-16n15.6
|
si:dkey-16n15.6 |
| chr21_-_31290582 | 0.09 |
ENSDART00000065362
|
ca4c
|
carbonic anhydrase IV c |
| chr15_-_31366742 | 0.09 |
ENSDART00000125585
|
or111-3
|
odorant receptor, family D, subfamily 111, member 3 |
| chr8_+_47188154 | 0.09 |
ENSDART00000137319
|
si:dkeyp-100a1.6
|
si:dkeyp-100a1.6 |
| chr13_-_50309969 | 0.09 |
ENSDART00000191597
|
CU928131.1
|
|
| chr5_+_37069605 | 0.09 |
ENSDART00000161977
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr9_+_48415043 | 0.08 |
ENSDART00000159930
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr14_-_25078569 | 0.08 |
ENSDART00000172802
ENSDART00000173345 ENSDART00000135004 |
matr3l1.1
|
matrin 3-like 1.1 |
| chr23_+_19790962 | 0.08 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr2_+_11670270 | 0.07 |
ENSDART00000100524
|
ftr01
|
finTRIM family, member 1 |
| chr22_+_19407531 | 0.07 |
ENSDART00000141060
|
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr6_+_27339962 | 0.06 |
ENSDART00000193726
|
klhl30
|
kelch-like family member 30 |
| chr9_-_44953664 | 0.06 |
ENSDART00000188558
ENSDART00000185210 |
vil1
|
villin 1 |
| chr19_+_1003332 | 0.06 |
ENSDART00000081779
|
zdhhc3b
|
zinc finger, DHHC-type containing 3b |
| chr24_+_1294176 | 0.06 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr13_+_18520738 | 0.05 |
ENSDART00000113952
|
tlr4al
|
toll-like receptor 4a, like |
| chr25_+_29472361 | 0.05 |
ENSDART00000154857
|
il17rel
|
interleukin 17 receptor E-like |
| chr6_+_52877147 | 0.05 |
ENSDART00000174370
|
or137-8
|
odorant receptor, family H, subfamily 137, member 8 |
| chr7_-_29341233 | 0.05 |
ENSDART00000140938
ENSDART00000147251 |
trpm1a
|
transient receptor potential cation channel, subfamily M, member 1a |
| chr9_+_1138323 | 0.05 |
ENSDART00000190352
ENSDART00000190387 |
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr22_-_20376488 | 0.05 |
ENSDART00000140187
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr5_-_65319387 | 0.04 |
ENSDART00000164649
|
col27a1b
|
collagen, type XXVII, alpha 1b |
| chr14_+_989733 | 0.04 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr11_+_30729745 | 0.04 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr1_-_57129179 | 0.04 |
ENSDART00000157226
ENSDART00000152469 |
si:ch73-94k4.2
|
si:ch73-94k4.2 |
| chr7_+_39738505 | 0.04 |
ENSDART00000004365
|
tada2b
|
transcriptional adaptor 2B |
| chr17_+_20174044 | 0.03 |
ENSDART00000156028
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr6_-_607063 | 0.03 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr4_+_38981587 | 0.02 |
ENSDART00000142713
|
si:dkey-66k12.3
|
si:dkey-66k12.3 |
| chr4_+_57099307 | 0.02 |
ENSDART00000131654
|
si:ch211-238e22.2
|
si:ch211-238e22.2 |
| chr15_-_31401361 | 0.02 |
ENSDART00000124360
|
or111-7
|
odorant receptor, family D, subfamily 111, member 7 |
| chr7_+_30823749 | 0.01 |
ENSDART00000085661
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr8_+_14778292 | 0.00 |
ENSDART00000089971
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr12_-_44122412 | 0.00 |
ENSDART00000169094
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hnf4g
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 23.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.4 | 1.3 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.3 | 2.4 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 0.7 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 1.7 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.2 | 1.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.7 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.8 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 1.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 1.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.5 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 1.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.5 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.1 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.6 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 1.5 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.2 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.9 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 1.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0021571 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.8 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.2 | GO:0043092 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.7 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 1.0 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 1.0 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.1 | GO:0070293 | renal absorption(GO:0070293) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 1.2 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.2 | 1.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.6 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 23.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.9 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 2.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.2 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 23.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.8 | 2.5 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.4 | 1.3 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.2 | 1.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 1.7 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.8 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 1.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 8.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 2.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.8 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.2 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0008329 | lipopolysaccharide receptor activity(GO:0001875) signaling pattern recognition receptor activity(GO:0008329) |
| 0.0 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 0.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |