Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
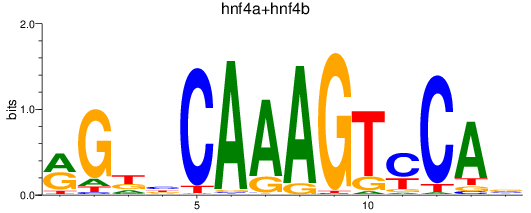
Results for hnf4a+hnf4b
Z-value: 1.37
Transcription factors associated with hnf4a+hnf4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hnf4a
|
ENSDARG00000021494 | hepatocyte nuclear factor 4, alpha |
|
hnf4b
|
ENSDARG00000104742 | hepatic nuclear factor 4, beta |
|
hnf4a
|
ENSDARG00000112985 | hepatocyte nuclear factor 4, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hnf4a | dr11_v1_chr23_+_25856541_25856641 | -0.79 | 8.2e-05 | Click! |
| CABZ01057488.1 | dr11_v1_chr7_+_69019851_69019851 | 0.67 | 2.3e-03 | Click! |
Activity profile of hnf4a+hnf4b motif
Sorted Z-values of hnf4a+hnf4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_2584423 | 5.54 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr20_+_54304800 | 5.29 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr19_-_27564458 | 4.46 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr9_+_8380728 | 4.45 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr20_-_43741159 | 4.34 |
ENSDART00000192621
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr16_-_17197546 | 4.33 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_+_24387659 | 4.18 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr23_+_2740741 | 4.17 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr17_-_2590222 | 4.13 |
ENSDART00000185711
|
CR759892.1
|
|
| chr17_-_2578026 | 4.06 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr17_-_2595736 | 3.98 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr19_-_27564980 | 3.96 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr16_-_17200120 | 3.94 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr17_-_2573021 | 3.83 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr7_-_24520866 | 3.31 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr16_-_25233515 | 3.09 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr22_-_24992532 | 2.94 |
ENSDART00000102751
|
si:dkey-179j5.5
|
si:dkey-179j5.5 |
| chr19_-_18130567 | 2.70 |
ENSDART00000190659
ENSDART00000022803 |
snx10a
|
sorting nexin 10a |
| chr3_-_36440705 | 2.31 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr7_-_20611039 | 2.27 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr25_+_8925934 | 2.21 |
ENSDART00000073914
|
accs
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) |
| chr18_-_46183462 | 2.18 |
ENSDART00000021192
|
kcnk6
|
potassium channel, subfamily K, member 6 |
| chr3_+_27770110 | 2.09 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr15_-_30857350 | 2.08 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr8_-_1264893 | 1.99 |
ENSDART00000190371
|
cdc14b
|
cell division cycle 14B |
| chr17_+_1360192 | 1.95 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr11_-_23687158 | 1.73 |
ENSDART00000189599
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr5_+_9218318 | 1.70 |
ENSDART00000137774
|
si:ch211-12e13.1
|
si:ch211-12e13.1 |
| chr7_+_24520518 | 1.66 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr5_-_36597612 | 1.66 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr14_+_24934736 | 1.59 |
ENSDART00000191821
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr24_+_35911020 | 1.59 |
ENSDART00000088480
|
abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr16_+_22345513 | 1.58 |
ENSDART00000078000
|
zgc:123238
|
zgc:123238 |
| chr15_+_29728377 | 1.57 |
ENSDART00000099958
|
zgc:153372
|
zgc:153372 |
| chr23_-_31810222 | 1.56 |
ENSDART00000134319
ENSDART00000139076 |
hbs1l
|
HBS1-like translational GTPase |
| chr23_+_9522942 | 1.55 |
ENSDART00000137751
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr6_+_40952031 | 1.55 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr24_+_35911300 | 1.54 |
ENSDART00000129679
|
abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr8_+_23827571 | 1.52 |
ENSDART00000040362
|
mapk14a
|
mitogen-activated protein kinase 14a |
| chr23_+_30730121 | 1.48 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr18_+_27515640 | 1.47 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr20_+_36806398 | 1.40 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr3_-_34561624 | 1.40 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr3_+_31662126 | 1.39 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr23_+_9522781 | 1.39 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr22_-_31517300 | 1.38 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr14_+_30413312 | 1.38 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr14_+_989733 | 1.37 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr7_+_25126629 | 1.37 |
ENSDART00000077217
|
zgc:101765
|
zgc:101765 |
| chr15_+_6459847 | 1.36 |
ENSDART00000157250
ENSDART00000065824 |
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr19_+_31532043 | 1.33 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr16_+_10264601 | 1.28 |
ENSDART00000186167
|
mrs2
|
MRS2 magnesium transporter |
| chr15_+_40074923 | 1.26 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr3_-_15470944 | 1.26 |
ENSDART00000185302
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr13_+_32807419 | 1.25 |
ENSDART00000085252
|
pqlc3
|
PQ loop repeat containing 3 |
| chr25_+_15997957 | 1.24 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr22_+_21549419 | 1.22 |
ENSDART00000139411
|
plpp2b
|
phospholipid phosphatase 2b |
| chr13_+_5013572 | 1.20 |
ENSDART00000162425
|
psap
|
prosaposin |
| chr15_-_20412286 | 1.20 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr17_-_49407091 | 1.20 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr11_-_18017287 | 1.19 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr6_-_41138854 | 1.18 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr18_-_12957451 | 1.18 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr15_+_34069746 | 1.18 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr23_-_43595956 | 1.16 |
ENSDART00000162186
|
itchb
|
itchy E3 ubiquitin protein ligase b |
| chr25_-_20049449 | 1.14 |
ENSDART00000104315
|
zgc:136858
|
zgc:136858 |
| chr1_-_12064715 | 1.14 |
ENSDART00000143628
ENSDART00000103406 |
pla2g12a
|
phospholipase A2, group XIIA |
| chr4_-_19693978 | 1.13 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr16_+_10264286 | 1.13 |
ENSDART00000091377
|
mrs2
|
MRS2 magnesium transporter |
| chr15_+_30310843 | 1.13 |
ENSDART00000112784
|
lyrm9
|
LYR motif containing 9 |
| chr10_+_8875195 | 1.05 |
ENSDART00000141045
|
itga2.3
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 3 |
| chr7_-_33868903 | 1.05 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr10_+_8197827 | 1.04 |
ENSDART00000026244
|
mtrex
|
Mtr4 exosome RNA helicase |
| chr5_+_32815745 | 1.01 |
ENSDART00000181535
|
crata
|
carnitine O-acetyltransferase a |
| chr22_-_7050 | 1.01 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr18_-_370286 | 1.01 |
ENSDART00000162633
|
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr13_+_8696825 | 1.01 |
ENSDART00000109059
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr13_-_35808904 | 0.99 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr17_+_33418475 | 0.98 |
ENSDART00000169145
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr23_-_31810391 | 0.96 |
ENSDART00000189749
ENSDART00000180850 |
hbs1l
|
HBS1-like translational GTPase |
| chr18_-_43866526 | 0.95 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr21_-_9446747 | 0.95 |
ENSDART00000158790
|
ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr2_-_9989919 | 0.92 |
ENSDART00000180213
ENSDART00000184369 |
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr5_-_55981288 | 0.90 |
ENSDART00000146616
|
si:dkey-189h5.6
|
si:dkey-189h5.6 |
| chr17_+_15216022 | 0.88 |
ENSDART00000138831
|
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr7_+_24115082 | 0.88 |
ENSDART00000182718
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr5_+_31959954 | 0.87 |
ENSDART00000142826
|
myo1hb
|
myosin IHb |
| chr14_+_24935131 | 0.84 |
ENSDART00000170871
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr18_-_43866001 | 0.83 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr21_-_11054876 | 0.81 |
ENSDART00000146576
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr23_-_3759345 | 0.81 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr22_-_16317886 | 0.78 |
ENSDART00000163664
|
trmt13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr21_+_41743493 | 0.76 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr8_-_9570511 | 0.76 |
ENSDART00000044000
|
plxna3
|
plexin A3 |
| chr25_+_418932 | 0.70 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr8_+_47099033 | 0.69 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr25_-_8625601 | 0.67 |
ENSDART00000155280
|
GDPGP1
|
zgc:153343 |
| chr7_-_51461649 | 0.66 |
ENSDART00000193947
ENSDART00000174328 |
arhgap36
|
Rho GTPase activating protein 36 |
| chr21_-_27272657 | 0.65 |
ENSDART00000040754
ENSDART00000175009 |
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr2_-_56649883 | 0.64 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr9_+_41690153 | 0.64 |
ENSDART00000100226
|
ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr22_-_10774735 | 0.64 |
ENSDART00000081156
|
timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr12_+_19138452 | 0.64 |
ENSDART00000141346
ENSDART00000066397 |
phf5a
|
PHD finger protein 5A |
| chr11_+_11152214 | 0.63 |
ENSDART00000148030
|
ly75
|
lymphocyte antigen 75 |
| chr3_-_50139860 | 0.62 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr4_+_11172964 | 0.62 |
ENSDART00000193598
ENSDART00000138661 |
tspan11
|
tetraspanin 11 |
| chr8_+_54137350 | 0.60 |
ENSDART00000164153
|
brpf1
|
bromodomain and PHD finger containing, 1 |
| chr5_-_41638039 | 0.60 |
ENSDART00000144525
|
epg5
|
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
| chr22_+_30502555 | 0.60 |
ENSDART00000139128
ENSDART00000104747 |
zgc:171679
|
zgc:171679 |
| chr7_+_65398161 | 0.59 |
ENSDART00000166109
ENSDART00000157399 |
usp47
|
ubiquitin specific peptidase 47 |
| chr14_+_30413758 | 0.57 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr2_+_6067371 | 0.56 |
ENSDART00000053868
ENSDART00000145244 |
aldh9a1b
|
aldehyde dehydrogenase 9 family, member A1b |
| chr9_+_8898024 | 0.55 |
ENSDART00000134954
ENSDART00000143671 ENSDART00000147098 ENSDART00000179969 ENSDART00000111214 ENSDART00000140232 ENSDART00000139687 ENSDART00000132443 ENSDART00000130208 |
naxd
|
NAD(P)HX dehydratase |
| chr21_-_13055195 | 0.55 |
ENSDART00000133517
|
myorg
|
myogenesis regulating glycosidase (putative) |
| chr16_+_7985886 | 0.55 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr8_-_20838342 | 0.54 |
ENSDART00000141345
|
si:ch211-133l5.7
|
si:ch211-133l5.7 |
| chr9_+_21383620 | 0.54 |
ENSDART00000130023
ENSDART00000062656 ENSDART00000180081 |
cryl1
|
crystallin, lambda 1 |
| chr10_+_10738880 | 0.54 |
ENSDART00000004181
|
slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr7_+_24114694 | 0.54 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr23_-_3759692 | 0.54 |
ENSDART00000028885
|
hmga1a
|
high mobility group AT-hook 1a |
| chr22_-_11648094 | 0.53 |
ENSDART00000191791
|
dpp4
|
dipeptidyl-peptidase 4 |
| chr7_-_60351876 | 0.53 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr5_-_13076779 | 0.53 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr20_+_27093042 | 0.52 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr8_+_6576940 | 0.52 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr20_+_39250673 | 0.52 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr23_-_44786844 | 0.51 |
ENSDART00000148669
|
si:ch73-269m23.5
|
si:ch73-269m23.5 |
| chr5_-_67267678 | 0.49 |
ENSDART00000156795
ENSDART00000125781 ENSDART00000125874 |
zcchc8
|
zinc finger, CCHC domain containing 8 |
| chr10_-_11385155 | 0.48 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr5_-_62317496 | 0.46 |
ENSDART00000180089
|
zgc:85789
|
zgc:85789 |
| chr18_+_30157034 | 0.46 |
ENSDART00000177669
|
gse1
|
Gse1 coiled-coil protein |
| chr18_+_30157198 | 0.46 |
ENSDART00000172579
|
gse1
|
Gse1 coiled-coil protein |
| chr8_-_65189 | 0.45 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr6_+_55819038 | 0.44 |
ENSDART00000108786
|
si:ch211-81n22.1
|
si:ch211-81n22.1 |
| chr2_-_22660232 | 0.43 |
ENSDART00000174742
|
thap4
|
THAP domain containing 4 |
| chr22_+_29994093 | 0.43 |
ENSDART00000104778
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr19_+_3215466 | 0.41 |
ENSDART00000181288
|
zgc:86598
|
zgc:86598 |
| chr2_-_22659450 | 0.40 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr10_+_45128375 | 0.40 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr4_+_6736746 | 0.38 |
ENSDART00000184175
|
tmem168a
|
transmembrane protein 168a |
| chr6_-_10912424 | 0.38 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr1_-_46862190 | 0.37 |
ENSDART00000145167
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr21_+_1119046 | 0.37 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr12_+_48681601 | 0.37 |
ENSDART00000187831
|
uros
|
uroporphyrinogen III synthase |
| chr10_-_6454386 | 0.36 |
ENSDART00000168549
|
ca9
|
carbonic anhydrase IX |
| chr23_-_21758253 | 0.35 |
ENSDART00000046613
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr15_+_15403560 | 0.34 |
ENSDART00000049831
|
dhrs11b
|
dehydrogenase/reductase (SDR family) member 11b |
| chr7_-_5487593 | 0.33 |
ENSDART00000136594
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr17_+_23926796 | 0.32 |
ENSDART00000021177
|
pex13
|
peroxisomal biogenesis factor 13 |
| chr5_-_67115872 | 0.32 |
ENSDART00000065262
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr14_+_6182346 | 0.30 |
ENSDART00000075525
|
si:ch73-22a13.3
|
si:ch73-22a13.3 |
| chr11_+_22134540 | 0.29 |
ENSDART00000190502
|
MDFI
|
si:dkey-91m3.1 |
| chr6_+_18367388 | 0.29 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr8_+_8699085 | 0.28 |
ENSDART00000021209
|
uxt
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr21_-_27273147 | 0.28 |
ENSDART00000143239
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr14_+_6429399 | 0.28 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr11_-_36957127 | 0.26 |
ENSDART00000168528
|
cacna1da
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, a |
| chr16_+_23924040 | 0.25 |
ENSDART00000161124
|
si:dkey-7f3.14
|
si:dkey-7f3.14 |
| chr6_+_36807861 | 0.25 |
ENSDART00000161708
|
si:ch73-29l19.1
|
si:ch73-29l19.1 |
| chr6_-_46474483 | 0.25 |
ENSDART00000155761
|
rdh20
|
retinol dehydrogenase 20 |
| chr21_-_18262287 | 0.24 |
ENSDART00000176716
|
vav2
|
vav 2 guanine nucleotide exchange factor |
| chr4_+_9360697 | 0.23 |
ENSDART00000128937
ENSDART00000156946 |
ipo8
|
importin 8 |
| chr23_-_27479558 | 0.23 |
ENSDART00000013563
|
atf7a
|
activating transcription factor 7a |
| chr15_-_1885247 | 0.23 |
ENSDART00000149703
|
porb
|
P450 (cytochrome) oxidoreductase b |
| chr16_+_23947196 | 0.22 |
ENSDART00000103190
ENSDART00000132961 ENSDART00000147690 ENSDART00000142168 |
apoa4b.2
|
apolipoprotein A-IV b, tandem duplicate 2 |
| chr20_-_25669813 | 0.22 |
ENSDART00000153118
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr18_+_31410652 | 0.21 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr6_-_43221581 | 0.21 |
ENSDART00000112239
|
si:dkey-178o16.4
|
si:dkey-178o16.4 |
| chr3_-_58116314 | 0.21 |
ENSDART00000154901
|
si:ch211-256e16.6
|
si:ch211-256e16.6 |
| chr11_-_30364847 | 0.20 |
ENSDART00000078387
|
pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr22_+_2417105 | 0.20 |
ENSDART00000106415
|
zgc:113220
|
zgc:113220 |
| chr3_+_12744083 | 0.20 |
ENSDART00000158554
ENSDART00000169545 |
cyp2k21
|
cytochrome P450, family 2, subfamily k, polypeptide 21 |
| chr9_-_1986014 | 0.20 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr7_+_52135791 | 0.20 |
ENSDART00000098705
|
cyp2x12
|
cytochrome P450, family 2, subfamily X, polypeptide 12 |
| chr2_-_24407933 | 0.19 |
ENSDART00000088584
|
si:dkey-208k22.6
|
si:dkey-208k22.6 |
| chr4_+_33547662 | 0.18 |
ENSDART00000150439
|
si:dkey-84h14.2
|
si:dkey-84h14.2 |
| chr6_-_35046735 | 0.18 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr12_-_44199316 | 0.18 |
ENSDART00000170378
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr8_+_1766206 | 0.18 |
ENSDART00000021820
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr11_+_30296332 | 0.18 |
ENSDART00000192843
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr11_+_14281100 | 0.17 |
ENSDART00000193664
ENSDART00000161597 |
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr24_+_19593197 | 0.17 |
ENSDART00000151923
|
slco5a1a
|
solute carrier organic anion transporter family member 5A1a |
| chr18_-_34143189 | 0.16 |
ENSDART00000079341
|
plch1
|
phospholipase C, eta 1 |
| chr7_+_31051603 | 0.16 |
ENSDART00000108721
|
tjp1a
|
tight junction protein 1a |
| chr13_+_12405051 | 0.16 |
ENSDART00000108543
|
atp10d
|
ATPase phospholipid transporting 10D |
| chr16_-_25607266 | 0.16 |
ENSDART00000192602
|
zgc:110410
|
zgc:110410 |
| chr6_-_43047774 | 0.16 |
ENSDART00000161722
|
glyctk
|
glycerate kinase |
| chr12_+_26877381 | 0.16 |
ENSDART00000087329
|
znf438
|
zinc finger protein 438 |
| chr24_-_36727922 | 0.15 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr25_-_27735290 | 0.14 |
ENSDART00000140663
|
zgc:153935
|
zgc:153935 |
| chr18_+_36037223 | 0.14 |
ENSDART00000144410
|
tmem91
|
transmembrane protein 91 |
| chr21_-_22910520 | 0.13 |
ENSDART00000065567
ENSDART00000191792 |
guca1d
|
guanylate cyclase activator 1d |
| chr23_-_27702561 | 0.13 |
ENSDART00000053876
|
dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr15_-_26552652 | 0.13 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr25_-_12803723 | 0.13 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr2_-_36914281 | 0.13 |
ENSDART00000008322
|
si:dkey-193b15.5
|
si:dkey-193b15.5 |
| chr20_-_46128590 | 0.12 |
ENSDART00000123744
|
taar1b
|
trace amine associated receptor 1b |
| chr20_-_40754794 | 0.12 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr16_-_25606889 | 0.12 |
ENSDART00000077447
ENSDART00000131528 |
zgc:110410
|
zgc:110410 |
| chr1_+_17900306 | 0.11 |
ENSDART00000089480
|
cyp4v8
|
cytochrome P450, family 4, subfamily V, polypeptide 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hnf4a+hnf4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 17.4 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.6 | 1.8 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.5 | 2.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.5 | 1.6 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.5 | 3.7 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.5 | 4.2 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.4 | 4.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.3 | 1.0 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 8.3 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.2 | 1.5 | GO:0040016 | embryonic cleavage(GO:0040016) positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 1.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 2.0 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 1.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 2.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.8 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 1.4 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 1.3 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 0.8 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 2.7 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 1.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.6 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 2.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.2 | GO:1903173 | fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 0.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.6 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 2.4 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 1.3 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 1.0 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 1.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 1.0 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.5 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.0 | 1.9 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.3 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.3 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 2.1 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0010459 | cardiac conduction system development(GO:0003161) negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 1.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 1.2 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 1.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.2 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 1.4 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.3 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.6 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.3 | 0.9 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.5 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 1.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.3 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 3.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 15.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.4 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0005689 | U2 snRNP(GO:0005686) U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 2.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 4.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.1 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0045178 | basal plasma membrane(GO:0009925) basal part of cell(GO:0045178) |
| 0.0 | 1.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.8 | GO:0010008 | endosome membrane(GO:0010008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.3 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.6 | 17.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 1.0 | 4.2 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.6 | 1.8 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.5 | 1.6 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.4 | 2.4 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.3 | 2.5 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.3 | 1.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 0.5 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.3 | 1.0 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.2 | 3.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 0.6 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.2 | 0.5 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.2 | 1.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.7 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 1.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.8 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.4 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.1 | 16.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 0.3 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 1.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 1.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 1.9 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 3.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 2.2 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 2.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 1.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.4 | 7.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.3 | 1.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 2.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 2.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |