Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
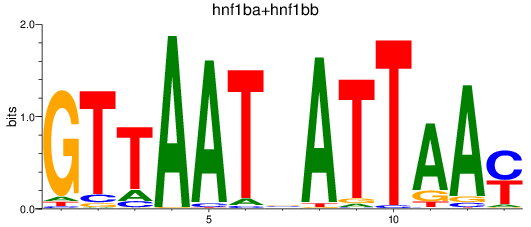
Results for hnf1ba+hnf1bb
Z-value: 1.16
Transcription factors associated with hnf1ba+hnf1bb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hnf1ba
|
ENSDARG00000006615 | HNF1 homeobox Ba |
|
hnf1bb
|
ENSDARG00000022295 | HNF1 homeobox Bb |
|
hnf1ba
|
ENSDARG00000115522 | HNF1 homeobox Ba |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hnf1bb | dr11_v1_chr21_+_38817785_38817843 | 0.40 | 9.9e-02 | Click! |
| hnf1ba | dr11_v1_chr15_-_16012963_16012963 | 0.37 | 1.3e-01 | Click! |
Activity profile of hnf1ba+hnf1bb motif
Sorted Z-values of hnf1ba+hnf1bb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_4765740 | 3.66 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr16_-_42894628 | 3.50 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr11_-_37997419 | 3.49 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr20_+_23440632 | 3.46 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr24_+_35787629 | 3.19 |
ENSDART00000136721
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr7_-_35066457 | 3.11 |
ENSDART00000058067
|
zgc:112160
|
zgc:112160 |
| chr24_-_39610585 | 3.09 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr21_+_38002879 | 2.99 |
ENSDART00000065183
|
cldn2
|
claudin 2 |
| chr20_-_31075972 | 2.85 |
ENSDART00000122927
|
si:ch211-198b3.4
|
si:ch211-198b3.4 |
| chr3_+_26734162 | 2.56 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr5_-_19394440 | 2.49 |
ENSDART00000163771
|
foxn4
|
forkhead box N4 |
| chr25_-_37191929 | 2.31 |
ENSDART00000128108
|
urahb
|
urate (5-hydroxyiso-) hydrolase b |
| chr2_-_17828279 | 2.31 |
ENSDART00000083314
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr5_-_30715225 | 2.28 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr3_-_16250527 | 2.14 |
ENSDART00000146699
ENSDART00000141181 |
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr24_+_26345427 | 2.13 |
ENSDART00000089756
|
lrrc34
|
leucine rich repeat containing 34 |
| chr18_-_48550426 | 2.02 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr4_+_7827261 | 1.95 |
ENSDART00000129568
|
phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr2_+_26647472 | 1.82 |
ENSDART00000145415
ENSDART00000157409 |
ttpa
|
tocopherol (alpha) transfer protein |
| chr25_-_35497055 | 1.69 |
ENSDART00000009271
|
slc5a12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr5_+_13385837 | 1.65 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr1_+_10051763 | 1.63 |
ENSDART00000011701
|
fgb
|
fibrinogen beta chain |
| chr24_+_26345609 | 1.54 |
ENSDART00000186844
|
lrrc34
|
leucine rich repeat containing 34 |
| chr1_-_19502322 | 1.54 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr7_-_25895189 | 1.51 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr8_+_26522013 | 1.45 |
ENSDART00000046863
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr13_-_36534380 | 1.43 |
ENSDART00000177857
ENSDART00000181046 ENSDART00000187243 |
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr24_+_23791758 | 1.40 |
ENSDART00000066655
ENSDART00000146580 |
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr1_+_34224360 | 1.21 |
ENSDART00000192938
|
arl6
|
ADP-ribosylation factor-like 6 |
| chr4_-_9533345 | 1.21 |
ENSDART00000128501
|
si:ch73-139j3.4
|
si:ch73-139j3.4 |
| chr18_+_3530769 | 1.16 |
ENSDART00000163469
|
si:ch73-338a16.3
|
si:ch73-338a16.3 |
| chr20_+_41756996 | 1.11 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr10_-_41664427 | 1.09 |
ENSDART00000150213
|
ggt1b
|
gamma-glutamyltransferase 1b |
| chr20_-_47347962 | 1.05 |
ENSDART00000080863
|
dtnba
|
dystrobrevin, beta a |
| chr25_+_23280220 | 1.04 |
ENSDART00000153940
|
ptprjb.1
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 1 |
| chr20_-_23440955 | 1.03 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr6_+_60055168 | 1.02 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr8_-_47800754 | 1.02 |
ENSDART00000115050
ENSDART00000133669 |
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr5_+_9428876 | 1.01 |
ENSDART00000081791
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr25_+_3347461 | 1.00 |
ENSDART00000104888
|
slc35b4
|
solute carrier family 35, member B4 |
| chr20_-_47348116 | 0.97 |
ENSDART00000162087
ENSDART00000160769 ENSDART00000164484 |
dtnba
|
dystrobrevin, beta a |
| chr7_-_72277269 | 0.90 |
ENSDART00000161914
ENSDART00000183927 |
slc35c1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr5_+_21225017 | 0.89 |
ENSDART00000141573
|
si:dkey-13n15.11
|
si:dkey-13n15.11 |
| chr12_+_35067863 | 0.88 |
ENSDART00000152849
|
syt15
|
synaptotagmin XV |
| chr5_+_45921234 | 0.86 |
ENSDART00000134355
|
ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr9_+_29994439 | 0.83 |
ENSDART00000012447
|
tmem30c
|
transmembrane protein 30C |
| chr5_+_37854685 | 0.82 |
ENSDART00000051222
ENSDART00000185283 |
ins
|
preproinsulin |
| chr24_-_6078222 | 0.81 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr15_+_28202170 | 0.81 |
ENSDART00000077736
|
vtna
|
vitronectin a |
| chr20_+_10538025 | 0.81 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr10_-_17159761 | 0.80 |
ENSDART00000080449
|
slc5a1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr3_+_13559199 | 0.80 |
ENSDART00000166547
|
si:ch73-106n3.1
|
si:ch73-106n3.1 |
| chr2_-_127945 | 0.75 |
ENSDART00000056453
|
igfbp1b
|
insulin-like growth factor binding protein 1b |
| chr20_+_10544100 | 0.74 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr22_-_23748284 | 0.73 |
ENSDART00000162005
|
cfhl2
|
complement factor H like 2 |
| chr5_+_32345187 | 0.73 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr13_-_23666579 | 0.73 |
ENSDART00000192457
ENSDART00000179777 |
map3k21
|
mitogen-activated protein kinase kinase kinase 21 |
| chr17_+_15033822 | 0.70 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr9_+_28688574 | 0.68 |
ENSDART00000101319
|
zgc:162396
|
zgc:162396 |
| chr7_+_27059330 | 0.67 |
ENSDART00000173919
|
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr1_-_22851481 | 0.62 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr5_+_45677781 | 0.62 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr14_+_38654300 | 0.61 |
ENSDART00000148938
ENSDART00000150107 |
rnasel3
|
ribonuclease like 3 |
| chr10_-_42131408 | 0.60 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr15_-_41744509 | 0.60 |
ENSDART00000176952
ENSDART00000156690 |
ftr88
|
finTRIM family, member 88 |
| chr8_+_15254564 | 0.59 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr22_-_23668356 | 0.59 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr10_+_25369254 | 0.58 |
ENSDART00000164375
|
bach1b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 b |
| chr3_-_9722603 | 0.57 |
ENSDART00000168234
|
crebbpb
|
CREB binding protein b |
| chr25_+_17920361 | 0.57 |
ENSDART00000185644
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr23_+_27740788 | 0.57 |
ENSDART00000053871
|
dhh
|
desert hedgehog |
| chr25_+_13406069 | 0.56 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr6_+_54221654 | 0.54 |
ENSDART00000128456
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr13_-_40726865 | 0.53 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr5_-_41307550 | 0.52 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr17_-_17948587 | 0.52 |
ENSDART00000090447
|
hhipl1
|
HHIP-like 1 |
| chr3_-_16760923 | 0.51 |
ENSDART00000055855
|
aspdh
|
aspartate dehydrogenase domain containing |
| chr3_+_40170216 | 0.51 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr6_+_41039166 | 0.50 |
ENSDART00000125659
|
entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr24_-_17039638 | 0.49 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr22_+_16555939 | 0.48 |
ENSDART00000012604
|
pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr6_+_7977611 | 0.48 |
ENSDART00000112290
|
palm3
|
paralemmin 3 |
| chr24_+_23716918 | 0.48 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr20_-_43723860 | 0.47 |
ENSDART00000122051
|
mixl1
|
Mix paired-like homeobox |
| chr5_-_12063381 | 0.46 |
ENSDART00000026749
|
nipsnap1
|
nipsnap homolog 1 (C. elegans) |
| chr12_-_3940768 | 0.46 |
ENSDART00000134292
|
zgc:92040
|
zgc:92040 |
| chr8_-_1051438 | 0.45 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr22_-_26005894 | 0.45 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr8_+_30699429 | 0.44 |
ENSDART00000005345
|
upb1
|
ureidopropionase, beta |
| chr11_+_575665 | 0.44 |
ENSDART00000122133
|
mkrn2os.1
|
MKRN2 opposite strand, tandem duplicate 1 |
| chr11_-_18601955 | 0.44 |
ENSDART00000180565
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr15_+_35043007 | 0.44 |
ENSDART00000086954
|
sesn3
|
sestrin 3 |
| chr6_-_59563597 | 0.42 |
ENSDART00000166311
|
INHBE
|
inhibin beta E subunit |
| chr10_+_21867307 | 0.42 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr5_-_42904329 | 0.41 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr5_+_28830643 | 0.39 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr7_+_27317174 | 0.39 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr4_+_45484774 | 0.37 |
ENSDART00000150573
|
si:dkey-256i11.6
|
si:dkey-256i11.6 |
| chr6_+_10094061 | 0.35 |
ENSDART00000150998
ENSDART00000162236 |
atp7b
|
ATPase copper transporting beta |
| chr24_+_14713776 | 0.35 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr8_-_53490376 | 0.34 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr4_+_18804317 | 0.34 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr21_-_23308286 | 0.33 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr8_+_26293673 | 0.33 |
ENSDART00000144977
|
mgll
|
monoglyceride lipase |
| chr20_-_40755614 | 0.33 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr22_-_26100282 | 0.33 |
ENSDART00000166075
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr1_+_22851261 | 0.32 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr6_+_28141135 | 0.32 |
ENSDART00000113405
ENSDART00000179795 |
inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr10_+_25369859 | 0.31 |
ENSDART00000047541
|
bach1b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 b |
| chr20_-_42534504 | 0.31 |
ENSDART00000132310
|
rfx6
|
regulatory factor X, 6 |
| chr6_+_30456788 | 0.31 |
ENSDART00000121492
|
FP236735.1
|
|
| chr22_+_17261801 | 0.31 |
ENSDART00000192978
ENSDART00000193187 ENSDART00000179953 ENSDART00000134798 |
tdrd5
|
tudor domain containing 5 |
| chr5_+_28830388 | 0.29 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr24_+_25913162 | 0.29 |
ENSDART00000143099
ENSDART00000184814 |
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr5_-_67661102 | 0.29 |
ENSDART00000013605
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr21_+_43669943 | 0.29 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr2_-_41942666 | 0.28 |
ENSDART00000075673
|
ebi3
|
Epstein-Barr virus induced 3 |
| chr4_-_5317006 | 0.28 |
ENSDART00000150867
|
si:ch211-214j24.15
|
si:ch211-214j24.15 |
| chr20_+_51006362 | 0.27 |
ENSDART00000028084
|
mpp5b
|
membrane protein, palmitoylated 5b (MAGUK p55 subfamily member 5) |
| chr23_+_27740592 | 0.26 |
ENSDART00000137875
|
dhh
|
desert hedgehog |
| chr7_-_59564011 | 0.26 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr17_-_51893123 | 0.25 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
| chr7_-_29534001 | 0.25 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr20_-_40750953 | 0.24 |
ENSDART00000061256
|
cx28.9
|
connexin 28.9 |
| chr12_+_5209822 | 0.23 |
ENSDART00000152610
|
SLC35G1
|
si:ch211-197g18.2 |
| chr13_+_23988442 | 0.23 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr2_-_27609189 | 0.20 |
ENSDART00000040555
|
lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr19_-_3876877 | 0.19 |
ENSDART00000163711
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr5_+_45322734 | 0.19 |
ENSDART00000084411
|
adamts3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr4_+_76416268 | 0.17 |
ENSDART00000174351
ENSDART00000191591 |
zgc:113209
FP074874.1
|
zgc:113209 |
| chr6_-_49078702 | 0.17 |
ENSDART00000135185
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr11_+_7432533 | 0.16 |
ENSDART00000180977
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr2_+_17055069 | 0.14 |
ENSDART00000115078
|
thpo
|
thrombopoietin |
| chr12_-_6177894 | 0.14 |
ENSDART00000152292
|
a1cf
|
apobec1 complementation factor |
| chr22_-_24818066 | 0.13 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr4_-_32642681 | 0.12 |
ENSDART00000188695
ENSDART00000193974 |
si:dkey-265e7.1
|
si:dkey-265e7.1 |
| chr6_+_28140816 | 0.12 |
ENSDART00000137907
|
inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr23_+_28770225 | 0.12 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr23_+_32335871 | 0.12 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr23_+_33752275 | 0.09 |
ENSDART00000007260
|
si:ch211-210c8.6
|
si:ch211-210c8.6 |
| chr2_+_1487118 | 0.09 |
ENSDART00000147283
|
c8a
|
complement component 8, alpha polypeptide |
| chr17_-_50233493 | 0.09 |
ENSDART00000172266
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr17_+_5895500 | 0.08 |
ENSDART00000019905
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr11_-_45152702 | 0.08 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr6_+_39930825 | 0.07 |
ENSDART00000182352
ENSDART00000186643 ENSDART00000190513 ENSDART00000065092 ENSDART00000188645 |
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr5_+_1877464 | 0.07 |
ENSDART00000050658
|
zgc:101699
|
zgc:101699 |
| chr7_+_23292133 | 0.06 |
ENSDART00000134489
|
htr2cl1
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 1 |
| chr2_+_1486822 | 0.06 |
ENSDART00000132500
|
c8a
|
complement component 8, alpha polypeptide |
| chr6_-_29515709 | 0.05 |
ENSDART00000180205
|
pex5la
|
peroxisomal biogenesis factor 5-like a |
| chr1_+_26667872 | 0.05 |
ENSDART00000152803
ENSDART00000152144 ENSDART00000152785 ENSDART00000152393 |
hemgn
|
hemogen |
| chr4_+_58016732 | 0.05 |
ENSDART00000165777
|
CT027609.1
|
|
| chr5_-_30620625 | 0.05 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr7_+_19817306 | 0.05 |
ENSDART00000044425
|
BX957278.1
|
|
| chr4_+_54798291 | 0.04 |
ENSDART00000165113
ENSDART00000109624 |
si:dkeyp-82b4.6
|
si:dkeyp-82b4.6 |
| chr4_-_32179699 | 0.04 |
ENSDART00000124106
ENSDART00000158835 |
si:dkey-72l17.6
|
si:dkey-72l17.6 |
| chr15_+_35139457 | 0.04 |
ENSDART00000059204
|
slc19a3a
|
solute carrier family 19 (thiamine transporter), member 3a |
| chr6_-_43028270 | 0.04 |
ENSDART00000149092
ENSDART00000149559 |
glyctk
|
glycerate kinase |
| chr12_+_27061720 | 0.03 |
ENSDART00000153426
|
srcap
|
Snf2-related CREBBP activator protein |
| chr19_+_19772765 | 0.03 |
ENSDART00000182028
ENSDART00000161019 |
hoxa3a
|
homeobox A3a |
| chr16_-_22294265 | 0.03 |
ENSDART00000124718
|
aqp10a
|
aquaporin 10a |
| chr7_-_17412559 | 0.02 |
ENSDART00000163020
|
nitr3a
|
novel immune-type receptor 3a |
| chr10_+_25726694 | 0.02 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr2_-_43635777 | 0.01 |
ENSDART00000148633
|
itgb1b.1
|
integrin, beta 1b.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hnf1ba+hnf1bb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.4 | 1.5 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.3 | 3.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.3 | 2.0 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 2.3 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.3 | 1.0 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.2 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.8 | GO:2000252 | positive regulation of epithelial cell differentiation(GO:0030858) negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.2 | 0.9 | GO:0072530 | purine-containing compound transmembrane transport(GO:0072530) |
| 0.2 | 0.8 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 1.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.8 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 1.6 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.3 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 1.8 | GO:0006775 | fat-soluble vitamin metabolic process(GO:0006775) |
| 0.1 | 2.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0044320 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.1 | 0.4 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 1.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.6 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.4 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 3.3 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.4 | GO:1902623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.4 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 1.0 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.3 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 1.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.5 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 3.7 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.7 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 1.2 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.3 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.5 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.5 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.2 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 2.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.0 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.9 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.7 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.6 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.3 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 3.2 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.0 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.8 | GO:0005770 | late endosome(GO:0005770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.2 | 1.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 1.0 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.2 | 0.5 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.2 | 1.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 3.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 2.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 1.1 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.4 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 1.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.8 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.7 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.9 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 1.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 2.2 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 1.8 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.0 | 0.8 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.5 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 1.0 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 2.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 1.8 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 3.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 3.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 3.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |