Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
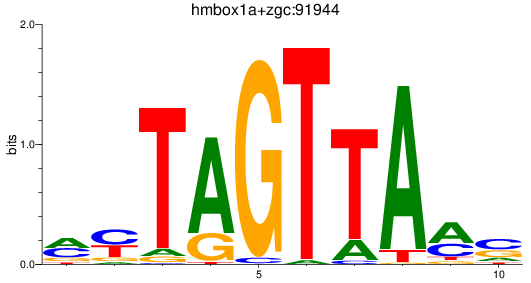
Results for hmbox1a+zgc:91944
Z-value: 0.40
Transcription factors associated with hmbox1a+zgc:91944
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmbox1a
|
ENSDARG00000027082 | homeobox containing 1a |
|
zgc
|
ENSDARG00000035887 | 91944 |
|
hmbox1a
|
ENSDARG00000109287 | homeobox containing 1 b |
|
zgc
|
ENSDARG00000114642 | 91944 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hmbox1a | dr11_v1_chr17_-_16324565_16324565 | 0.84 | 1.6e-05 | Click! |
| CABZ01078261.1 | dr11_v1_chr20_+_49787584_49787584 | 0.75 | 3.4e-04 | Click! |
| zgc:91944 | dr11_v1_chr19_-_32600823_32600868 | -0.30 | 2.3e-01 | Click! |
Activity profile of hmbox1a+zgc:91944 motif
Sorted Z-values of hmbox1a+zgc:91944 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_35191220 | 0.63 |
ENSDART00000110552
|
zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr18_+_35130416 | 0.59 |
ENSDART00000151595
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr2_-_21438492 | 0.54 |
ENSDART00000046098
|
plcd1b
|
phospholipase C, delta 1b |
| chr10_+_7709724 | 0.52 |
ENSDART00000097670
|
ggcx
|
gamma-glutamyl carboxylase |
| chr17_+_21902058 | 0.52 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr13_-_21672131 | 0.46 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr2_-_27619954 | 0.45 |
ENSDART00000144826
|
tgs1
|
trimethylguanosine synthase 1 |
| chr7_+_36467796 | 0.43 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr15_-_1036878 | 0.41 |
ENSDART00000123844
|
si:dkey-77f5.3
|
si:dkey-77f5.3 |
| chr17_-_11368662 | 0.38 |
ENSDART00000159061
ENSDART00000188694 ENSDART00000190932 |
si:ch211-185a18.2
|
si:ch211-185a18.2 |
| chr15_+_6459847 | 0.37 |
ENSDART00000157250
ENSDART00000065824 |
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr19_-_20430892 | 0.37 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr13_+_39182099 | 0.36 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr19_+_43669122 | 0.35 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr23_+_25868637 | 0.35 |
ENSDART00000103934
|
ttpal
|
tocopherol (alpha) transfer protein-like |
| chr3_-_23512285 | 0.34 |
ENSDART00000159151
|
BX682558.1
|
|
| chr23_-_36857964 | 0.34 |
ENSDART00000188822
ENSDART00000134061 ENSDART00000093061 |
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr5_-_11809404 | 0.33 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr4_+_26056548 | 0.33 |
ENSDART00000171204
|
SCYL2
|
si:ch211-244b2.1 |
| chr12_-_13650344 | 0.32 |
ENSDART00000124364
ENSDART00000124638 ENSDART00000171929 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr17_-_15188440 | 0.31 |
ENSDART00000151885
|
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr10_-_26196383 | 0.31 |
ENSDART00000192925
|
fhdc3
|
FH2 domain containing 3 |
| chr16_-_42965192 | 0.31 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr7_-_25133783 | 0.31 |
ENSDART00000173781
ENSDART00000121943 ENSDART00000077219 |
badb
|
BCL2 associated agonist of cell death b |
| chr3_-_26806032 | 0.30 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr2_+_23701613 | 0.30 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr12_+_19281189 | 0.30 |
ENSDART00000153428
|
tnrc6b
|
trinucleotide repeat containing 6b |
| chr13_-_36050303 | 0.29 |
ENSDART00000134955
ENSDART00000139087 |
lgmn
|
legumain |
| chr12_+_10706772 | 0.29 |
ENSDART00000158227
|
top2a
|
DNA topoisomerase II alpha |
| chr6_+_44197348 | 0.29 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr11_+_25129013 | 0.29 |
ENSDART00000132879
|
ndrg3a
|
ndrg family member 3a |
| chr22_-_1079773 | 0.29 |
ENSDART00000136668
|
si:ch1073-15f12.3
|
si:ch1073-15f12.3 |
| chr19_-_20099394 | 0.28 |
ENSDART00000126173
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr10_-_26202766 | 0.28 |
ENSDART00000136393
|
fhdc3
|
FH2 domain containing 3 |
| chr1_-_23596391 | 0.28 |
ENSDART00000155184
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr18_-_20458840 | 0.27 |
ENSDART00000177125
|
kif23
|
kinesin family member 23 |
| chrM_+_6425 | 0.27 |
ENSDART00000093606
|
mt-co1
|
cytochrome c oxidase I, mitochondrial |
| chr21_-_36453417 | 0.27 |
ENSDART00000018350
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr12_-_26538823 | 0.27 |
ENSDART00000143213
|
acsf2
|
acyl-CoA synthetase family member 2 |
| chr9_-_54248182 | 0.26 |
ENSDART00000129540
|
larsa
|
leucyl-tRNA synthetase a |
| chr9_-_9228941 | 0.26 |
ENSDART00000121665
|
cbsb
|
cystathionine-beta-synthase b |
| chr10_-_44008241 | 0.26 |
ENSDART00000137686
|
acads
|
acyl-CoA dehydrogenase short chain |
| chr20_-_33512275 | 0.26 |
ENSDART00000185959
|
paplnb
|
papilin b, proteoglycan-like sulfated glycoprotein |
| chr2_+_23677179 | 0.26 |
ENSDART00000153918
|
oxsr1a
|
oxidative stress responsive 1a |
| chr3_-_47128096 | 0.25 |
ENSDART00000063287
|
qtrt1
|
queuine tRNA-ribosyltransferase 1 |
| chr6_+_13506841 | 0.25 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr10_+_16036246 | 0.25 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr21_+_37090585 | 0.24 |
ENSDART00000182971
|
znf346
|
zinc finger protein 346 |
| chr16_-_17699111 | 0.24 |
ENSDART00000108581
|
si:dkey-17m8.1
|
si:dkey-17m8.1 |
| chr23_+_13528053 | 0.24 |
ENSDART00000162217
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr10_+_41939963 | 0.24 |
ENSDART00000126248
|
tmem120b
|
transmembrane protein 120B |
| chr9_+_29431763 | 0.23 |
ENSDART00000186095
ENSDART00000182640 |
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr24_-_7587401 | 0.23 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr16_+_33992418 | 0.23 |
ENSDART00000101885
ENSDART00000130540 |
zdhhc18a
|
zinc finger, DHHC-type containing 18a |
| chr16_+_33121106 | 0.22 |
ENSDART00000110195
|
akirin1
|
akirin 1 |
| chr1_-_11104805 | 0.22 |
ENSDART00000147648
|
knl1
|
kinetochore scaffold 1 |
| chr3_+_30922947 | 0.22 |
ENSDART00000184060
|
cldni
|
claudin i |
| chr20_-_34670236 | 0.22 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr4_+_2059692 | 0.22 |
ENSDART00000067427
|
yeats4
|
YEATS domain containing 4 |
| chr11_+_42474694 | 0.21 |
ENSDART00000056048
ENSDART00000184710 |
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr5_-_15283509 | 0.21 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr10_-_22912255 | 0.21 |
ENSDART00000131992
|
si:ch1073-143l10.2
|
si:ch1073-143l10.2 |
| chr11_+_25157374 | 0.20 |
ENSDART00000019450
|
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr18_+_13077800 | 0.20 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr13_+_28785814 | 0.20 |
ENSDART00000039028
|
nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr12_-_18577774 | 0.20 |
ENSDART00000078166
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr25_+_28825657 | 0.20 |
ENSDART00000153625
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr1_+_513986 | 0.20 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr3_+_13842554 | 0.20 |
ENSDART00000162317
ENSDART00000158068 |
ilf3b
|
interleukin enhancer binding factor 3b |
| chr22_-_36770649 | 0.20 |
ENSDART00000169442
|
acy1
|
aminoacylase 1 |
| chr13_+_46718518 | 0.20 |
ENSDART00000160401
ENSDART00000182884 |
tmem63ba
|
transmembrane protein 63Ba |
| chr12_-_49151326 | 0.19 |
ENSDART00000153244
|
bub3
|
BUB3 mitotic checkpoint protein |
| chr10_+_45302425 | 0.19 |
ENSDART00000159954
|
zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr13_+_8840772 | 0.19 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr7_+_13609457 | 0.19 |
ENSDART00000172857
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr24_+_28953089 | 0.19 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr3_+_22335030 | 0.19 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr12_-_27588299 | 0.19 |
ENSDART00000178023
ENSDART00000066282 |
dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr11_-_6868287 | 0.19 |
ENSDART00000037824
|
lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr13_-_24906307 | 0.19 |
ENSDART00000148191
ENSDART00000189810 |
kat6b
|
K(lysine) acetyltransferase 6B |
| chr18_-_31051847 | 0.19 |
ENSDART00000170982
|
gas8
|
growth arrest-specific 8 |
| chr7_+_17938128 | 0.18 |
ENSDART00000141044
|
mta2
|
metastasis associated 1 family, member 2 |
| chr25_-_25736958 | 0.18 |
ENSDART00000166308
|
cib2
|
calcium and integrin binding family member 2 |
| chr17_+_30591287 | 0.18 |
ENSDART00000154243
|
si:dkey-190l8.2
|
si:dkey-190l8.2 |
| chr5_+_20257225 | 0.18 |
ENSDART00000127919
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr12_-_999762 | 0.18 |
ENSDART00000127003
ENSDART00000084076 ENSDART00000152425 |
mettl9
|
methyltransferase like 9 |
| chr16_+_29690708 | 0.17 |
ENSDART00000103054
|
lysmd1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr6_-_40657653 | 0.17 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr9_-_25328527 | 0.17 |
ENSDART00000060840
|
med4
|
mediator complex subunit 4 |
| chr13_+_48359573 | 0.17 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr22_+_28818291 | 0.17 |
ENSDART00000136032
|
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr7_-_38570878 | 0.17 |
ENSDART00000139187
ENSDART00000134570 ENSDART00000041055 |
celf1
|
cugbp, Elav-like family member 1 |
| chr25_-_19486399 | 0.17 |
ENSDART00000155076
ENSDART00000156016 |
zgc:193812
|
zgc:193812 |
| chr15_+_19335646 | 0.16 |
ENSDART00000062569
|
acad8
|
acyl-CoA dehydrogenase family, member 8 |
| chr12_+_3723650 | 0.16 |
ENSDART00000179922
ENSDART00000152482 ENSDART00000108771 |
pagr1
|
PAXIP1 associated glutamate-rich protein 1 |
| chr19_+_7559901 | 0.16 |
ENSDART00000141189
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr21_-_2042037 | 0.16 |
ENSDART00000171131
ENSDART00000160144 |
add1
|
adducin 1 (alpha) |
| chr2_-_32366287 | 0.16 |
ENSDART00000144758
|
ubtfl
|
upstream binding transcription factor, like |
| chr3_+_33367954 | 0.16 |
ENSDART00000103161
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr8_-_28259766 | 0.16 |
ENSDART00000134226
ENSDART00000087037 ENSDART00000136973 |
ddx20
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 |
| chr14_+_23717165 | 0.16 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr17_+_4325693 | 0.15 |
ENSDART00000154264
|
mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr1_-_23110740 | 0.15 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr3_-_31254379 | 0.15 |
ENSDART00000189376
|
apnl
|
actinoporin-like protein |
| chr23_-_17101360 | 0.15 |
ENSDART00000053418
|
dnmt3bb.1
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.1 |
| chr4_-_2059233 | 0.14 |
ENSDART00000188177
ENSDART00000129521 ENSDART00000082289 |
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr14_+_23709134 | 0.14 |
ENSDART00000191162
ENSDART00000179754 ENSDART00000054266 |
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr10_-_28513861 | 0.14 |
ENSDART00000177781
|
bbx
|
bobby sox homolog (Drosophila) |
| chr2_+_3403264 | 0.13 |
ENSDART00000105387
|
jmjd4
|
jumonji domain containing 4 |
| chr17_-_7818944 | 0.13 |
ENSDART00000135538
ENSDART00000037541 |
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr10_+_22912251 | 0.13 |
ENSDART00000134790
|
med11
|
mediator complex subunit 11 |
| chr13_+_9612395 | 0.13 |
ENSDART00000136689
ENSDART00000138362 |
slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr7_-_23848012 | 0.13 |
ENSDART00000146104
ENSDART00000175108 |
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr5_+_25733774 | 0.13 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr2_-_24061575 | 0.13 |
ENSDART00000089234
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr21_-_32036597 | 0.13 |
ENSDART00000114964
|
zgc:165573
|
zgc:165573 |
| chr4_+_9011448 | 0.13 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr2_-_57264262 | 0.12 |
ENSDART00000183815
ENSDART00000149829 ENSDART00000088508 ENSDART00000149508 |
mbd3a
|
methyl-CpG binding domain protein 3a |
| chr19_+_2602903 | 0.12 |
ENSDART00000033132
|
fam126a
|
family with sequence similarity 126, member A |
| chr8_-_47152001 | 0.12 |
ENSDART00000163922
ENSDART00000110512 ENSDART00000024320 |
ybx1
|
Y box binding protein 1 |
| chr23_-_18982499 | 0.12 |
ENSDART00000012507
|
bcl2l1
|
bcl2-like 1 |
| chr9_-_49805218 | 0.12 |
ENSDART00000179290
|
GALNT3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr3_+_28831622 | 0.11 |
ENSDART00000184130
ENSDART00000191294 |
flr
|
fleer |
| chr18_-_44331041 | 0.11 |
ENSDART00000168619
|
prdm10
|
PR domain containing 10 |
| chr22_+_18315490 | 0.11 |
ENSDART00000160413
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr3_-_30186296 | 0.11 |
ENSDART00000134395
ENSDART00000077057 ENSDART00000017422 |
tbc1d17
|
TBC1 domain family, member 17 |
| chr1_+_30723380 | 0.11 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr12_-_33359052 | 0.11 |
ENSDART00000135943
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr2_-_14390627 | 0.11 |
ENSDART00000172367
|
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr2_+_33926606 | 0.11 |
ENSDART00000111430
|
kif2c
|
kinesin family member 2C |
| chr17_-_37184655 | 0.11 |
ENSDART00000180447
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr14_-_36437249 | 0.10 |
ENSDART00000016728
|
aga
|
aspartylglucosaminidase |
| chr11_+_11030908 | 0.10 |
ENSDART00000111091
|
pla2r1
|
phospholipase A2 receptor 1 |
| chr21_+_33459524 | 0.10 |
ENSDART00000053205
|
cd74b
|
CD74 molecule, major histocompatibility complex, class II invariant chain b |
| chr18_+_34601023 | 0.10 |
ENSDART00000149647
|
tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr6_-_37441520 | 0.10 |
ENSDART00000132586
|
nit2
|
nitrilase family, member 2 |
| chr16_+_54829574 | 0.10 |
ENSDART00000148392
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr2_+_30960351 | 0.10 |
ENSDART00000141575
|
lpin2
|
lipin 2 |
| chr25_+_25737386 | 0.10 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr18_-_2727764 | 0.10 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr8_-_36112278 | 0.10 |
ENSDART00000183283
|
CT583723.1
|
|
| chr16_+_43401005 | 0.10 |
ENSDART00000110994
|
sqlea
|
squalene epoxidase a |
| chr12_+_35011899 | 0.09 |
ENSDART00000153007
ENSDART00000153020 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr8_+_17868506 | 0.09 |
ENSDART00000175436
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr7_-_41420226 | 0.09 |
ENSDART00000074155
|
abcf2b
|
ATP-binding cassette, sub-family F (GCN20), member 2b |
| chr15_+_577698 | 0.09 |
ENSDART00000155799
|
si:ch73-144d13.4
|
si:ch73-144d13.4 |
| chr20_-_47051996 | 0.09 |
ENSDART00000153330
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr1_+_44395976 | 0.09 |
ENSDART00000159686
ENSDART00000189905 ENSDART00000025145 |
unc93b1
|
unc-93 homolog B1, TLR signaling regulator |
| chr6_+_11990733 | 0.09 |
ENSDART00000151075
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr12_-_19153177 | 0.09 |
ENSDART00000057125
|
tefa
|
thyrotrophic embryonic factor a |
| chr1_-_29061285 | 0.08 |
ENSDART00000053933
ENSDART00000142350 ENSDART00000192615 |
gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr16_-_42004544 | 0.08 |
ENSDART00000034544
|
caspa
|
caspase a |
| chr9_-_55772937 | 0.08 |
ENSDART00000159192
|
akap17a
|
A kinase (PRKA) anchor protein 17A |
| chr9_-_32753535 | 0.08 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr24_-_18809433 | 0.08 |
ENSDART00000152009
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr9_-_1483879 | 0.08 |
ENSDART00000093415
|
rbm45
|
RNA binding motif protein 45 |
| chr9_+_29643036 | 0.08 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr4_+_72798545 | 0.08 |
ENSDART00000181727
|
MYRFL
|
myelin regulatory factor-like |
| chr4_-_14486822 | 0.08 |
ENSDART00000048821
|
plxnb2a
|
plexin b2a |
| chr2_-_43191465 | 0.08 |
ENSDART00000025254
|
crema
|
cAMP responsive element modulator a |
| chr6_-_32314913 | 0.07 |
ENSDART00000051779
|
atg4c
|
autophagy related 4C, cysteine peptidase |
| chr23_-_30960506 | 0.07 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr19_+_32201991 | 0.07 |
ENSDART00000022667
|
fam8a1a
|
family with sequence similarity 8, member A1a |
| chr3_-_11878490 | 0.07 |
ENSDART00000129961
|
coro7
|
coronin 7 |
| chr11_+_43043171 | 0.07 |
ENSDART00000180344
|
CABZ01092982.1
|
|
| chr15_-_25518084 | 0.07 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr6_-_31325400 | 0.07 |
ENSDART00000188869
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr18_-_6881392 | 0.07 |
ENSDART00000154968
|
si:dkey-266m15.7
|
si:dkey-266m15.7 |
| chr4_-_77506362 | 0.06 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr5_+_19448078 | 0.06 |
ENSDART00000088968
|
ube3b
|
ubiquitin protein ligase E3B |
| chr10_-_11012000 | 0.06 |
ENSDART00000132995
|
ak3
|
adenylate kinase 3 |
| chr21_+_43404945 | 0.06 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr20_+_32552912 | 0.06 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr3_+_46764022 | 0.06 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr17_+_5985933 | 0.06 |
ENSDART00000190844
|
zgc:194275
|
zgc:194275 |
| chr17_+_28624321 | 0.06 |
ENSDART00000122260
|
heatr5a
|
HEAT repeat containing 5a |
| chr3_+_54761569 | 0.06 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr19_+_31585917 | 0.05 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr17_-_40876680 | 0.05 |
ENSDART00000127200
|
supt7l
|
SPT7-like STAGA complex gamma subunit |
| chr1_-_9641845 | 0.05 |
ENSDART00000121490
ENSDART00000159411 |
ugt5b2
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B2 UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr13_-_24429066 | 0.05 |
ENSDART00000147514
ENSDART00000057588 |
cep68
|
centrosomal protein 68 |
| chr11_-_29650930 | 0.05 |
ENSDART00000166969
|
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr5_-_29559758 | 0.04 |
ENSDART00000051471
ENSDART00000167115 |
nelfb
|
negative elongation factor complex member B |
| chr9_+_37850360 | 0.04 |
ENSDART00000185148
ENSDART00000082165 ENSDART00000133643 |
sec22a
|
SEC22 homolog A, vesicle trafficking protein |
| chr8_+_10339869 | 0.04 |
ENSDART00000132253
|
tbc1d22b
|
TBC1 domain family, member 22B |
| chr24_-_20321012 | 0.04 |
ENSDART00000163683
|
map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr16_+_26738404 | 0.04 |
ENSDART00000163110
|
fsbp
|
fibrinogen silencer binding protein |
| chr17_-_14451718 | 0.04 |
ENSDART00000039438
|
jkamp
|
jnk1/mapk8 associated membrane protein |
| chr3_+_32443395 | 0.04 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr1_-_51719110 | 0.04 |
ENSDART00000190574
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr22_-_16755885 | 0.04 |
ENSDART00000036467
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr18_+_5454341 | 0.04 |
ENSDART00000192649
|
dtwd1
|
DTW domain containing 1 |
| chr4_-_20313810 | 0.04 |
ENSDART00000136350
|
dcp1b
|
decapping mRNA 1B |
| chr19_+_31585341 | 0.04 |
ENSDART00000052185
|
gmnn
|
geminin, DNA replication inhibitor |
| chr6_-_48473694 | 0.04 |
ENSDART00000154237
|
ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr5_+_8246603 | 0.03 |
ENSDART00000167373
|
si:ch1073-475a24.1
|
si:ch1073-475a24.1 |
| chr24_-_4782052 | 0.03 |
ENSDART00000149911
|
agtr1b
|
angiotensin II receptor, type 1b |
| chr2_-_43123949 | 0.03 |
ENSDART00000075347
ENSDART00000132346 |
lrrc6
|
leucine rich repeat containing 6 |
| chr17_-_25382367 | 0.03 |
ENSDART00000162306
ENSDART00000165282 |
lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr8_+_14890821 | 0.03 |
ENSDART00000190966
|
soat1
|
sterol O-acyltransferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hmbox1a+zgc:91944
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.3 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 | 0.3 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.3 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 0.2 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.2 | GO:0071674 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of lamellipodium organization(GO:1902745) |
| 0.1 | 0.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.2 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0032239 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0019079 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0060907 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.4 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 1.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.4 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.2 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.2 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.2 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.2 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.0 | 0.3 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) positive regulation of mRNA catabolic process(GO:0061014) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.2 | GO:0044279 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.3 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.3 | GO:0031956 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.3 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.3 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |