Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
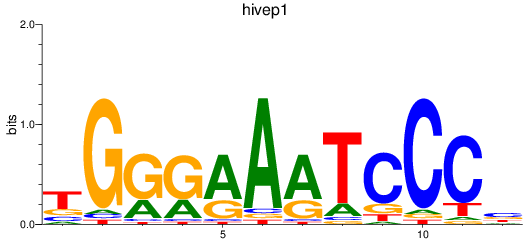
Results for hivep1
Z-value: 0.34
Transcription factors associated with hivep1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hivep1
|
ENSDARG00000103658 | HIVEP zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hivep1 | dr11_v1_chr19_-_3488860_3488860 | 0.34 | 1.7e-01 | Click! |
Activity profile of hivep1 motif
Sorted Z-values of hivep1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_43583896 | 0.60 |
ENSDART00000161711
|
itgb1b
|
integrin, beta 1b |
| chr19_-_18136410 | 0.48 |
ENSDART00000012352
|
cbx3a
|
chromobox homolog 3a (HP1 gamma homolog, Drosophila) |
| chr10_+_5954787 | 0.47 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr3_+_31662126 | 0.47 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr13_+_15816573 | 0.46 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr16_+_23403602 | 0.42 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr12_+_28888975 | 0.41 |
ENSDART00000076362
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr13_+_33368140 | 0.41 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr16_+_10557504 | 0.39 |
ENSDART00000091241
|
si:ch73-22o12.1
|
si:ch73-22o12.1 |
| chr1_+_292545 | 0.39 |
ENSDART00000148261
|
cenpe
|
centromere protein E |
| chr18_+_16053455 | 0.35 |
ENSDART00000189163
ENSDART00000188269 |
FO834850.1
|
|
| chr8_+_10823069 | 0.35 |
ENSDART00000081341
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr6_-_19333947 | 0.33 |
ENSDART00000160887
|
gga3
|
ggolgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr14_+_8947282 | 0.33 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr18_+_11858397 | 0.33 |
ENSDART00000133762
|
tmtc2b
|
transmembrane and tetratricopeptide repeat containing 2b |
| chr8_-_20230802 | 0.33 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr5_-_31689796 | 0.32 |
ENSDART00000184319
ENSDART00000190229 ENSDART00000186294 ENSDART00000170313 ENSDART00000147065 ENSDART00000134427 ENSDART00000098172 |
sh3glb2b
|
SH3-domain GRB2-like endophilin B2b |
| chr3_-_25055795 | 0.31 |
ENSDART00000156459
|
ep300b
|
E1A binding protein p300 b |
| chr5_-_29195063 | 0.31 |
ENSDART00000109926
|
man1b1b
|
mannosidase, alpha, class 1B, member 1b |
| chr8_-_20230559 | 0.30 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr23_-_21748805 | 0.30 |
ENSDART00000148128
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr6_+_32355204 | 0.29 |
ENSDART00000157878
|
dock7
|
dedicator of cytokinesis 7 |
| chr20_-_41992878 | 0.29 |
ENSDART00000100967
|
si:dkeyp-114g9.1
|
si:dkeyp-114g9.1 |
| chr25_+_16646113 | 0.29 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr6_+_2195625 | 0.29 |
ENSDART00000155659
|
acvr1bb
|
activin A receptor type 1Bb |
| chr20_+_23501535 | 0.28 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr10_+_37173029 | 0.27 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr6_+_54576520 | 0.27 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr18_-_21725638 | 0.27 |
ENSDART00000089853
|
fan1
|
FANCD2/FANCI-associated nuclease 1 |
| chr6_+_21227621 | 0.26 |
ENSDART00000193583
|
prkca
|
protein kinase C, alpha |
| chr13_+_22717939 | 0.26 |
ENSDART00000188288
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr14_+_22467672 | 0.26 |
ENSDART00000079409
ENSDART00000136597 |
nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr20_+_43691208 | 0.26 |
ENSDART00000152976
ENSDART00000045185 |
lin9
|
lin-9 DREAM MuvB core complex component |
| chr14_-_31087830 | 0.25 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr9_+_38588081 | 0.24 |
ENSDART00000031127
ENSDART00000131784 |
snx4
|
sorting nexin 4 |
| chr13_-_25196758 | 0.23 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr13_-_25819825 | 0.23 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr14_+_49152341 | 0.23 |
ENSDART00000084114
|
nsd1a
|
nuclear receptor binding SET domain protein 1a |
| chr3_+_61221194 | 0.23 |
ENSDART00000110030
|
lmtk2
|
lemur tyrosine kinase 2 |
| chr19_+_7549854 | 0.22 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr5_+_22579975 | 0.21 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr11_+_25064519 | 0.21 |
ENSDART00000016181
|
ndrg3a
|
ndrg family member 3a |
| chr17_-_13070602 | 0.21 |
ENSDART00000188311
ENSDART00000193428 |
CU469462.1
|
|
| chr12_+_31494830 | 0.21 |
ENSDART00000153390
|
gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr15_+_17100412 | 0.20 |
ENSDART00000154418
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr10_+_29963518 | 0.20 |
ENSDART00000011317
ENSDART00000099964 ENSDART00000182990 ENSDART00000113912 |
ntm
|
neurotrimin |
| chr15_+_17100697 | 0.20 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr20_+_13883131 | 0.20 |
ENSDART00000003248
ENSDART00000152611 |
nek2
|
NIMA-related kinase 2 |
| chr11_+_12719944 | 0.19 |
ENSDART00000054837
ENSDART00000144954 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr25_+_35889102 | 0.19 |
ENSDART00000023453
ENSDART00000125821 ENSDART00000135441 |
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr2_-_2642476 | 0.19 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr16_+_48631412 | 0.19 |
ENSDART00000154273
|
pbx2
|
pre-B-cell leukemia homeobox 2 |
| chr11_+_12720171 | 0.19 |
ENSDART00000135761
ENSDART00000122812 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr14_-_36345175 | 0.19 |
ENSDART00000077823
|
lrit3a
|
info leucine-rich repeat, immunoglobulin-like and transmembrane domains 3a |
| chr18_+_30567945 | 0.19 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr2_+_30547018 | 0.18 |
ENSDART00000193747
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr10_+_39199547 | 0.18 |
ENSDART00000075943
|
ei24
|
etoposide induced 2.4 |
| chr1_+_41498188 | 0.17 |
ENSDART00000191934
ENSDART00000146310 |
dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr18_+_16717764 | 0.17 |
ENSDART00000166849
|
eif4g2b
|
eukaryotic translation initiation factor 4, gamma 2b |
| chr20_+_13883353 | 0.17 |
ENSDART00000188006
|
nek2
|
NIMA-related kinase 2 |
| chr15_-_41677689 | 0.17 |
ENSDART00000187063
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr22_-_38951128 | 0.16 |
ENSDART00000085685
ENSDART00000104422 |
rbbp9
|
retinoblastoma binding protein 9 |
| chr10_+_5422575 | 0.15 |
ENSDART00000063093
|
auh
|
AU RNA binding protein/enoyl-CoA hydratase |
| chr3_+_32681274 | 0.15 |
ENSDART00000140290
ENSDART00000164428 |
kat8
|
K(lysine) acetyltransferase 8 |
| chr9_+_35014513 | 0.15 |
ENSDART00000100701
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr3_+_35406395 | 0.15 |
ENSDART00000055266
ENSDART00000150918 |
rbbp6
|
retinoblastoma binding protein 6 |
| chr9_+_35014728 | 0.14 |
ENSDART00000100700
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr20_-_44055095 | 0.14 |
ENSDART00000125898
ENSDART00000082265 |
runx2b
|
runt-related transcription factor 2b |
| chr15_-_16076399 | 0.14 |
ENSDART00000135658
ENSDART00000133755 ENSDART00000080413 |
srsf1a
|
serine/arginine-rich splicing factor 1a |
| chr1_+_594736 | 0.14 |
ENSDART00000166731
|
jam2a
|
junctional adhesion molecule 2a |
| chr2_-_37684641 | 0.13 |
ENSDART00000012191
|
hiat1a
|
hippocampus abundant transcript 1a |
| chr8_-_12432604 | 0.12 |
ENSDART00000133350
ENSDART00000140699 ENSDART00000101174 |
traf1
|
TNF receptor-associated factor 1 |
| chr7_-_7398350 | 0.12 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr9_+_34151367 | 0.12 |
ENSDART00000143991
|
gpr161
|
G protein-coupled receptor 161 |
| chr2_-_37401600 | 0.12 |
ENSDART00000015723
|
prkci
|
protein kinase C, iota |
| chr6_+_27146671 | 0.11 |
ENSDART00000156792
|
kif1aa
|
kinesin family member 1Aa |
| chr15_-_20412286 | 0.11 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr12_-_25217217 | 0.11 |
ENSDART00000152931
|
kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr14_-_16807206 | 0.11 |
ENSDART00000157957
|
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr8_+_52377516 | 0.11 |
ENSDART00000115398
|
arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr25_-_14637660 | 0.11 |
ENSDART00000143666
|
nav2b
|
neuron navigator 2b |
| chr5_+_37032038 | 0.10 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr3_-_1146497 | 0.10 |
ENSDART00000149061
|
si:ch73-211l13.2
|
si:ch73-211l13.2 |
| chr12_+_2381213 | 0.10 |
ENSDART00000188007
|
LO018238.1
|
|
| chr7_+_72276511 | 0.10 |
ENSDART00000166544
|
tollip
|
toll interacting protein |
| chr3_-_26904774 | 0.09 |
ENSDART00000103690
|
clec16a
|
C-type lectin domain containing 16A |
| chr18_+_10784730 | 0.09 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr22_+_8313513 | 0.09 |
ENSDART00000181169
ENSDART00000103911 |
CABZ01077217.1
|
|
| chr17_+_8183393 | 0.09 |
ENSDART00000155957
|
tulp4b
|
tubby like protein 4b |
| chr10_+_15088534 | 0.09 |
ENSDART00000142865
|
si:ch211-95j8.3
|
si:ch211-95j8.3 |
| chr19_+_40250682 | 0.08 |
ENSDART00000102888
|
cdk6
|
cyclin-dependent kinase 6 |
| chr1_-_18585046 | 0.08 |
ENSDART00000147228
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr1_+_46579885 | 0.08 |
ENSDART00000144821
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr19_-_1871415 | 0.08 |
ENSDART00000004585
|
clptm1l
|
CLPTM1-like |
| chr18_+_16715864 | 0.08 |
ENSDART00000079758
|
eif4g2b
|
eukaryotic translation initiation factor 4, gamma 2b |
| chr6_-_51541488 | 0.08 |
ENSDART00000156336
|
si:dkey-6e2.2
|
si:dkey-6e2.2 |
| chr3_+_56574623 | 0.08 |
ENSDART00000130877
|
rac1b
|
Rac family small GTPase 1b |
| chr4_+_8016457 | 0.08 |
ENSDART00000014036
|
optn
|
optineurin |
| chr19_+_7717449 | 0.07 |
ENSDART00000104719
ENSDART00000146747 |
tuft1b
|
tuftelin 1b |
| chr13_-_4664403 | 0.07 |
ENSDART00000023803
ENSDART00000177957 |
c1d
|
C1D nuclear receptor corepressor |
| chr5_-_52964789 | 0.07 |
ENSDART00000166267
|
zfpl1
|
zinc finger protein-like 1 |
| chr13_+_31177934 | 0.07 |
ENSDART00000144568
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr1_-_52447364 | 0.07 |
ENSDART00000140740
|
si:ch211-217k17.10
|
si:ch211-217k17.10 |
| chr9_-_51563575 | 0.07 |
ENSDART00000167034
ENSDART00000148918 |
tank
|
TRAF family member-associated NFKB activator |
| chr5_-_38107741 | 0.07 |
ENSDART00000156853
|
si:ch211-284e13.14
|
si:ch211-284e13.14 |
| chr25_-_36020344 | 0.07 |
ENSDART00000181448
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr9_-_32142311 | 0.06 |
ENSDART00000142768
|
ankrd44
|
ankyrin repeat domain 44 |
| chr22_+_9922301 | 0.06 |
ENSDART00000105924
|
blf
|
bloody fingers |
| chr2_+_111919 | 0.06 |
ENSDART00000149391
|
fggy
|
FGGY carbohydrate kinase domain containing |
| chr11_-_279328 | 0.06 |
ENSDART00000066179
|
npffl
|
neuropeptide FF-amide peptide precursor like |
| chr2_-_53592532 | 0.06 |
ENSDART00000184066
|
ccl25a
|
chemokine (C-C motif) ligand 25a |
| chr22_-_38951354 | 0.06 |
ENSDART00000104423
ENSDART00000134117 |
rbbp9
|
retinoblastoma binding protein 9 |
| chr20_-_51267185 | 0.06 |
ENSDART00000158350
ENSDART00000023064 |
slc35b2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr12_-_4835378 | 0.05 |
ENSDART00000172093
|
si:ch211-93e11.8
|
si:ch211-93e11.8 |
| chr20_-_51307815 | 0.05 |
ENSDART00000098833
|
nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr23_-_33738570 | 0.05 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr7_-_6680622 | 0.05 |
ENSDART00000180437
ENSDART00000173270 ENSDART00000066450 |
dusp19b
|
dual specificity phosphatase 19b |
| chr19_-_11208782 | 0.05 |
ENSDART00000044426
ENSDART00000189754 |
si:dkey-240h12.4
|
si:dkey-240h12.4 |
| chr2_+_25868404 | 0.05 |
ENSDART00000143517
ENSDART00000187743 |
slc7a14a
|
solute carrier family 7, member 14a |
| chr18_-_40905901 | 0.05 |
ENSDART00000064848
|
pglyrp5
|
peptidoglycan recognition protein 5 |
| chr18_-_5527050 | 0.05 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr23_-_36823932 | 0.05 |
ENSDART00000142305
|
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr2_-_37465517 | 0.05 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr6_-_30859656 | 0.04 |
ENSDART00000156235
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr4_+_74943111 | 0.04 |
ENSDART00000004739
|
nup50
|
nucleoporin 50 |
| chr7_-_60831082 | 0.04 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr11_+_19370447 | 0.04 |
ENSDART00000186154
|
prickle2b
|
prickle homolog 2b |
| chr4_+_20263097 | 0.03 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr20_-_49657134 | 0.03 |
ENSDART00000151248
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr3_+_6325895 | 0.03 |
ENSDART00000167386
ENSDART00000165000 |
zgc:136767
|
zgc:136767 |
| chr14_-_22108718 | 0.03 |
ENSDART00000054410
|
med19a
|
mediator complex subunit 19a |
| chr21_-_2621127 | 0.03 |
ENSDART00000172363
|
col4a3bpb
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein b |
| chr5_+_70271799 | 0.02 |
ENSDART00000101316
|
znf618
|
zinc finger protein 618 |
| chr18_+_27738349 | 0.02 |
ENSDART00000187816
|
tspan18b
|
tetraspanin 18b |
| chr9_-_49978335 | 0.02 |
ENSDART00000170814
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr8_+_25247245 | 0.02 |
ENSDART00000045798
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr9_+_32859967 | 0.02 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr11_-_36020005 | 0.02 |
ENSDART00000031993
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr21_+_21791343 | 0.02 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr4_-_12914163 | 0.02 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr18_-_43880020 | 0.01 |
ENSDART00000185638
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr13_+_40692804 | 0.01 |
ENSDART00000109822
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr7_-_17591007 | 0.01 |
ENSDART00000171023
|
CU672228.1
|
|
| chr8_-_10949847 | 0.01 |
ENSDART00000123209
|
pqlc2
|
PQ loop repeat containing 2 |
| chr24_+_25919809 | 0.01 |
ENSDART00000006615
|
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr5_+_28830388 | 0.01 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr9_-_32663496 | 0.01 |
ENSDART00000078384
|
gzm3.2
|
granzyme 3, tandem duplicate 2 |
| chr23_+_19606291 | 0.01 |
ENSDART00000139415
|
flnb
|
filamin B, beta (actin binding protein 278) |
| chr24_-_38816725 | 0.01 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr8_+_17529151 | 0.01 |
ENSDART00000147760
|
si:ch73-70k4.1
|
si:ch73-70k4.1 |
| chr25_+_3347461 | 0.01 |
ENSDART00000104888
|
slc35b4
|
solute carrier family 35, member B4 |
| chr14_+_1014109 | 0.01 |
ENSDART00000157945
|
f8
|
coagulation factor VIII, procoagulant component |
| chr12_-_3903886 | 0.00 |
ENSDART00000184214
ENSDART00000041082 |
gdpd3b
|
glycerophosphodiester phosphodiesterase domain containing 3b |
| chr3_+_22273123 | 0.00 |
ENSDART00000044157
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr14_-_24081929 | 0.00 |
ENSDART00000158576
|
msx2a
|
muscle segment homeobox 2a |
| chr7_-_34262080 | 0.00 |
ENSDART00000183246
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hivep1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.2 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.0 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.6 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.3 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.0 | 0.4 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.4 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.3 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.4 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.0 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |