Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for hinfp
Z-value: 0.31
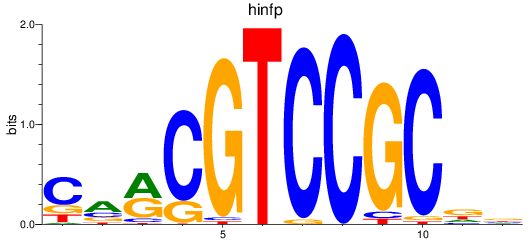
Transcription factors associated with hinfp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hinfp
|
ENSDARG00000004851 | histone H4 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hinfp | dr11_v1_chr5_+_30596477_30596477 | 0.70 | 1.3e-03 | Click! |
Activity profile of hinfp motif
Sorted Z-values of hinfp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_21362071 | 0.91 |
ENSDART00000125167
|
avd
|
avidin |
| chr9_+_8408054 | 0.88 |
ENSDART00000144373
|
zgc:153499
|
zgc:153499 |
| chr10_-_21362320 | 0.86 |
ENSDART00000189789
|
avd
|
avidin |
| chr5_-_63065912 | 0.80 |
ENSDART00000161149
|
fam57a
|
family with sequence similarity 57 member A |
| chr4_-_20081621 | 0.75 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr5_+_47863153 | 0.74 |
ENSDART00000051518
|
rasa1a
|
RAS p21 protein activator (GTPase activating protein) 1a |
| chr24_+_5799764 | 0.72 |
ENSDART00000154482
|
si:ch211-157j23.2
|
si:ch211-157j23.2 |
| chr5_+_47862636 | 0.68 |
ENSDART00000139824
|
rasa1a
|
RAS p21 protein activator (GTPase activating protein) 1a |
| chr9_+_8407778 | 0.68 |
ENSDART00000102754
ENSDART00000178144 |
zgc:153499
|
zgc:153499 |
| chr19_-_31765615 | 0.68 |
ENSDART00000103636
|
si:dkeyp-120h9.1
|
si:dkeyp-120h9.1 |
| chr5_-_47975440 | 0.65 |
ENSDART00000145665
ENSDART00000007057 |
ccnh
|
cyclin H |
| chr8_+_11687254 | 0.61 |
ENSDART00000042040
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr8_+_11687586 | 0.61 |
ENSDART00000146241
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr17_-_24866727 | 0.57 |
ENSDART00000027957
|
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr11_+_2710530 | 0.56 |
ENSDART00000132768
ENSDART00000030921 ENSDART00000040147 |
mapk14b
|
mitogen-activated protein kinase 14b |
| chr17_-_24866964 | 0.55 |
ENSDART00000190601
ENSDART00000192547 |
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr7_+_29163762 | 0.54 |
ENSDART00000173762
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr21_-_12749501 | 0.54 |
ENSDART00000179724
|
LO018011.1
|
|
| chr17_+_6793001 | 0.51 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr2_-_27651674 | 0.50 |
ENSDART00000177402
|
tgs1
|
trimethylguanosine synthase 1 |
| chr4_-_2196798 | 0.48 |
ENSDART00000110178
ENSDART00000149330 |
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr13_+_28618086 | 0.44 |
ENSDART00000087001
|
cnnm2a
|
cyclin and CBS domain divalent metal cation transport mediator 2a |
| chr5_+_31168096 | 0.42 |
ENSDART00000086443
ENSDART00000192271 |
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr2_+_15048410 | 0.41 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr20_+_42246948 | 0.41 |
ENSDART00000061135
|
gopc
|
golgi-associated PDZ and coiled-coil motif containing |
| chr16_+_23495600 | 0.41 |
ENSDART00000021092
|
snx27b
|
sorting nexin family member 27b |
| chr8_+_21225064 | 0.40 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr23_+_45845423 | 0.39 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr25_-_21763750 | 0.39 |
ENSDART00000089596
|
tmem168b
|
transmembrane protein 168b |
| chr7_-_69184420 | 0.38 |
ENSDART00000168311
ENSDART00000159239 ENSDART00000161319 |
usp10
|
ubiquitin specific peptidase 10 |
| chr20_+_28266892 | 0.36 |
ENSDART00000103330
|
chac1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr21_+_21279159 | 0.36 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr3_+_25166805 | 0.36 |
ENSDART00000077493
|
TST
|
zgc:162544 |
| chr3_+_26035550 | 0.35 |
ENSDART00000078137
|
ankrd54
|
ankyrin repeat domain 54 |
| chr7_-_48396193 | 0.34 |
ENSDART00000083555
|
sin3ab
|
SIN3 transcription regulator family member Ab |
| chr2_-_56635744 | 0.33 |
ENSDART00000167790
ENSDART00000168160 |
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr7_-_8961941 | 0.33 |
ENSDART00000111002
|
si:ch211-74f19.2
|
si:ch211-74f19.2 |
| chr16_-_17586883 | 0.32 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr21_+_28747069 | 0.32 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr4_-_20232974 | 0.31 |
ENSDART00000193353
|
stk38l
|
serine/threonine kinase 38 like |
| chr20_-_43917647 | 0.29 |
ENSDART00000026213
|
mark3b
|
MAP/microtubule affinity-regulating kinase 3b |
| chr19_+_8612839 | 0.29 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr2_+_27651984 | 0.29 |
ENSDART00000126762
|
tmem68
|
transmembrane protein 68 |
| chr20_+_35208020 | 0.29 |
ENSDART00000153315
ENSDART00000045135 |
fbxo16
|
F-box protein 16 |
| chr23_+_1216215 | 0.28 |
ENSDART00000165957
|
utrn
|
utrophin |
| chr11_-_21363834 | 0.28 |
ENSDART00000080051
|
RASSF5
|
si:dkey-85p17.3 |
| chr11_-_25853212 | 0.27 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr13_+_29925397 | 0.26 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr21_+_28747236 | 0.26 |
ENSDART00000137874
|
zgc:100829
|
zgc:100829 |
| chr2_-_1468258 | 0.26 |
ENSDART00000114431
|
gpaa1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chr23_+_43668756 | 0.25 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr23_+_30707837 | 0.24 |
ENSDART00000016096
|
dnajc11a
|
DnaJ (Hsp40) homolog, subfamily C, member 11a |
| chr10_+_21444654 | 0.24 |
ENSDART00000140113
ENSDART00000184386 ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr7_-_62244744 | 0.24 |
ENSDART00000192522
ENSDART00000170635 ENSDART00000073872 |
CCKAR
|
cholecystokinin A receptor |
| chr2_+_27652386 | 0.24 |
ENSDART00000188261
|
tmem68
|
transmembrane protein 68 |
| chr22_-_20342260 | 0.23 |
ENSDART00000161610
ENSDART00000165667 |
tcf3b
|
transcription factor 3b |
| chr5_-_23117078 | 0.22 |
ENSDART00000051529
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr5_-_37881345 | 0.22 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr1_+_57347888 | 0.21 |
ENSDART00000104222
|
b3gntl1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
| chr6_+_34038963 | 0.21 |
ENSDART00000057732
ENSDART00000192496 |
ap1m3
|
adaptor-related protein complex 1, mu 3 subunit |
| chr4_-_77557279 | 0.21 |
ENSDART00000180113
|
AL935186.10
|
|
| chr8_-_49935358 | 0.20 |
ENSDART00000159782
ENSDART00000156841 |
agtpbp1
CU694202.1
|
ATP/GTP binding protein 1 |
| chr25_-_31907590 | 0.20 |
ENSDART00000149707
ENSDART00000149471 |
eif3ja
|
eukaryotic translation initiation factor 3, subunit Ja |
| chr21_+_18950575 | 0.16 |
ENSDART00000036172
|
crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr17_-_21784152 | 0.15 |
ENSDART00000127254
|
hmx2
|
H6 family homeobox 2 |
| chr10_-_35321625 | 0.15 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr3_-_8399774 | 0.14 |
ENSDART00000190238
ENSDART00000113140 ENSDART00000132949 |
rbfox3b
|
RNA binding fox-1 homolog 3b |
| chr24_+_28383278 | 0.14 |
ENSDART00000018095
|
sh3glb1a
|
SH3-domain GRB2-like endophilin B1a |
| chr7_+_28612671 | 0.14 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr10_+_8551897 | 0.14 |
ENSDART00000084353
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr25_-_10791437 | 0.13 |
ENSDART00000127054
|
BX572619.1
|
|
| chr5_+_38886499 | 0.11 |
ENSDART00000076845
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr16_+_2905150 | 0.11 |
ENSDART00000109980
|
lars2
|
leucyl-tRNA synthetase 2, mitochondrial |
| chr23_+_1078072 | 0.11 |
ENSDART00000159263
ENSDART00000053527 |
slc34a2b
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 2b |
| chr5_-_8483457 | 0.10 |
ENSDART00000171886
ENSDART00000159477 ENSDART00000157423 |
lifra
|
leukemia inhibitory factor receptor alpha a |
| chr3_+_25849560 | 0.10 |
ENSDART00000007119
|
mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr3_-_6441619 | 0.10 |
ENSDART00000157771
ENSDART00000166758 |
sumo2b
|
small ubiquitin-like modifier 2b |
| chr19_-_31900893 | 0.10 |
ENSDART00000113797
|
zbtb10
|
zinc finger and BTB domain containing 10 |
| chr1_+_54069450 | 0.09 |
ENSDART00000108601
ENSDART00000187878 |
dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr6_-_58975010 | 0.09 |
ENSDART00000144911
ENSDART00000144514 |
mars
|
methionyl-tRNA synthetase |
| chr19_-_11425542 | 0.09 |
ENSDART00000177875
ENSDART00000080762 |
sept7a
|
septin 7a |
| chr5_-_56943064 | 0.09 |
ENSDART00000146991
|
si:ch211-127d4.3
|
si:ch211-127d4.3 |
| chr25_-_26833100 | 0.08 |
ENSDART00000014052
|
neil1
|
nei-like DNA glycosylase 1 |
| chr22_+_11775269 | 0.08 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr16_+_40954481 | 0.08 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr2_-_3678029 | 0.08 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr3_+_60962410 | 0.06 |
ENSDART00000042674
|
helz
|
helicase with zinc finger |
| chr11_+_1551603 | 0.06 |
ENSDART00000185383
ENSDART00000121489 ENSDART00000040577 |
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr12_-_34758474 | 0.06 |
ENSDART00000153418
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr10_+_13279079 | 0.06 |
ENSDART00000135082
|
tmem267
|
transmembrane protein 267 |
| chr6_-_35310224 | 0.06 |
ENSDART00000148997
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr21_-_1742159 | 0.05 |
ENSDART00000151049
|
onecut2
|
one cut homeobox 2 |
| chr21_-_10446405 | 0.05 |
ENSDART00000167948
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr8_+_41533268 | 0.05 |
ENSDART00000142377
|
si:ch211-158d24.2
|
si:ch211-158d24.2 |
| chr19_-_1023051 | 0.05 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr21_-_3770636 | 0.05 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr21_-_41147818 | 0.04 |
ENSDART00000167339
ENSDART00000192730 |
msx2b
|
muscle segment homeobox 2b |
| chr20_-_2298970 | 0.04 |
ENSDART00000136067
|
EPB41L2
|
si:ch73-18b11.1 |
| chr20_-_49650293 | 0.04 |
ENSDART00000151239
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr2_+_16173999 | 0.04 |
ENSDART00000177639
ENSDART00000157601 ENSDART00000190186 ENSDART00000045933 |
sh3glb1b
|
SH3-domain GRB2-like endophilin B1b |
| chr19_+_48290786 | 0.04 |
ENSDART00000165986
|
zgc:101785
|
zgc:101785 |
| chr22_-_24900121 | 0.04 |
ENSDART00000147643
|
si:dkey-4c23.3
|
si:dkey-4c23.3 |
| chr14_+_16937997 | 0.03 |
ENSDART00000163013
ENSDART00000167856 |
limch1b
|
LIM and calponin homology domains 1b |
| chr6_-_57655030 | 0.03 |
ENSDART00000155438
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr4_+_72580821 | 0.02 |
ENSDART00000124528
|
si:cabz01054394.5
|
si:cabz01054394.5 |
| chr12_+_46608361 | 0.02 |
ENSDART00000189164
|
FADS6
|
fatty acid desaturase 6 |
| chr1_+_40801448 | 0.02 |
ENSDART00000187394
|
cpz
|
carboxypeptidase Z |
| chr5_+_12888260 | 0.02 |
ENSDART00000175916
|
gal3st1a
|
galactose-3-O-sulfotransferase 1a |
| chr4_+_11690923 | 0.02 |
ENSDART00000150624
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr9_-_4856767 | 0.02 |
ENSDART00000016814
ENSDART00000138015 |
fmnl2a
|
formin-like 2a |
| chr20_-_6278817 | 0.02 |
ENSDART00000033404
|
opn7a
|
opsin 7, group member a |
| chr17_+_5846202 | 0.02 |
ENSDART00000189713
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr19_+_5135113 | 0.01 |
ENSDART00000151310
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr9_+_55154414 | 0.01 |
ENSDART00000182924
|
ANOS1
|
anosmin 1 |
| chr19_-_5135345 | 0.01 |
ENSDART00000151787
|
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr22_-_20105969 | 0.01 |
ENSDART00000088687
|
rxfp3.2b
|
relaxin/insulin-like family peptide receptor 3.2b |
| chr1_-_34612613 | 0.00 |
ENSDART00000004361
|
klf5a
|
Kruppel-like factor 5a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hinfp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.4 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.7 | GO:0033238 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.1 | 0.6 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.2 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 1.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.4 | GO:0010955 | glutathione catabolic process(GO:0006751) negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.1 | GO:0044321 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.0 | 0.4 | GO:0030330 | DNA damage response, signal transduction by p53 class mediator(GO:0030330) |
| 0.0 | 1.1 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.8 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.6 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 1.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 1.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |