Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
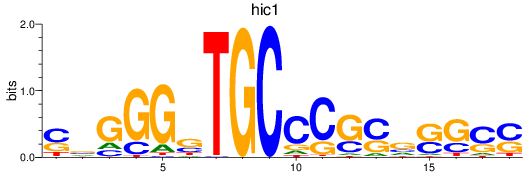
Results for hic1
Z-value: 0.27
Transcription factors associated with hic1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hic1
|
ENSDARG00000055493 | hypermethylated in cancer 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hic1 | dr11_v1_chr15_+_25683069_25683069 | 0.63 | 5.1e-03 | Click! |
Activity profile of hic1 motif
Sorted Z-values of hic1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_20916251 | 0.87 |
ENSDART00000134053
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr3_-_61592417 | 0.78 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr5_+_62723233 | 0.77 |
ENSDART00000183718
|
nanos2
|
nanos homolog 2 |
| chr15_-_18138607 | 0.59 |
ENSDART00000176690
|
CR385077.1
|
|
| chr13_-_37519774 | 0.52 |
ENSDART00000141420
ENSDART00000185478 |
sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr8_+_35172594 | 0.49 |
ENSDART00000177146
|
BX897670.1
|
|
| chr21_-_45878872 | 0.48 |
ENSDART00000029763
|
sap30l
|
sap30-like |
| chr16_+_50006145 | 0.45 |
ENSDART00000049375
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr1_+_135903 | 0.41 |
ENSDART00000124837
|
f10
|
coagulation factor X |
| chr17_-_8976307 | 0.28 |
ENSDART00000092113
|
zranb1b
|
zinc finger, RAN-binding domain containing 1b |
| chr14_+_49602 | 0.22 |
ENSDART00000035581
|
OTOP1
|
otopetrin 1 |
| chr5_-_52784152 | 0.21 |
ENSDART00000169307
|
fam189a2
|
family with sequence similarity 189, member A2 |
| chr14_-_21064199 | 0.19 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr4_-_55728559 | 0.16 |
ENSDART00000186201
|
CT583728.14
|
|
| chr14_-_21063977 | 0.15 |
ENSDART00000164373
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr4_+_90048 | 0.14 |
ENSDART00000166440
|
lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr1_-_59571758 | 0.14 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr10_+_23088 | 0.14 |
ENSDART00000022840
|
riox2
|
ribosomal oxygenase 2 |
| chr4_+_55778679 | 0.13 |
ENSDART00000183009
|
CT583728.19
|
|
| chr4_+_55794876 | 0.13 |
ENSDART00000189043
|
CT583728.17
|
|
| chr4_+_55810436 | 0.13 |
ENSDART00000182875
|
CT583728.16
|
|
| chr14_+_52369262 | 0.13 |
ENSDART00000169352
ENSDART00000157833 |
igfbp7
|
insulin-like growth factor binding protein 7 |
| chr4_-_68568233 | 0.12 |
ENSDART00000184284
|
BX548011.1
|
|
| chr4_-_797831 | 0.11 |
ENSDART00000158970
ENSDART00000170012 |
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr1_+_45839927 | 0.10 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr7_-_24995631 | 0.08 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr4_-_68831753 | 0.08 |
ENSDART00000189170
|
si:dkey-264f17.1
|
si:dkey-264f17.1 |
| chr3_-_56896702 | 0.07 |
ENSDART00000023265
|
ush1ga
|
Usher syndrome 1Ga (autosomal recessive) |
| chr14_-_34044369 | 0.06 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr19_-_19505167 | 0.06 |
ENSDART00000160582
|
creb5a
|
cAMP responsive element binding protein 5a |
| chr23_+_45611649 | 0.06 |
ENSDART00000169521
|
dclk2b
|
doublecortin-like kinase 2b |
| chr9_-_849069 | 0.05 |
ENSDART00000136091
|
c1ql2
|
complement component 1, q subcomponent-like 2 |
| chr23_+_14590767 | 0.05 |
ENSDART00000143675
|
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr1_+_10720294 | 0.04 |
ENSDART00000139387
|
atp1b1b
|
ATPase Na+/K+ transporting subunit beta 1b |
| chr10_-_21484065 | 0.04 |
ENSDART00000064545
|
fgf18b
|
fibroblast growth factor 18b |
| chr9_-_14992730 | 0.03 |
ENSDART00000137117
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr20_-_18915376 | 0.02 |
ENSDART00000063725
|
xkr6b
|
XK, Kell blood group complex subunit-related family, member 6b |
| chr9_+_7358749 | 0.02 |
ENSDART00000081660
|
ihha
|
Indian hedgehog homolog a |
| chr6_-_1874664 | 0.01 |
ENSDART00000007972
|
dlgap4b
|
discs, large (Drosophila) homolog-associated protein 4b |
| chr12_-_54375 | 0.01 |
ENSDART00000152304
|
si:ch1073-357b18.4
|
si:ch1073-357b18.4 |
| chr12_-_979789 | 0.01 |
ENSDART00000128188
|
daglb
|
diacylglycerol lipase, beta |
| chr7_-_61845282 | 0.01 |
ENSDART00000182586
|
HTRA3
|
HtrA serine peptidase 3 |
| chr1_+_9004719 | 0.00 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hic1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.3 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.5 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.8 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |