Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
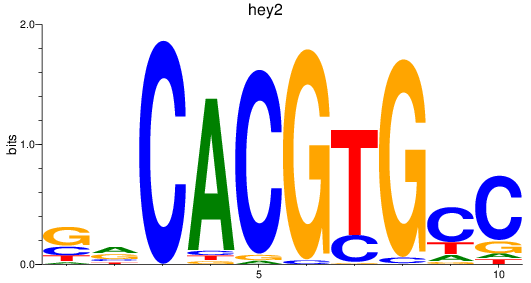
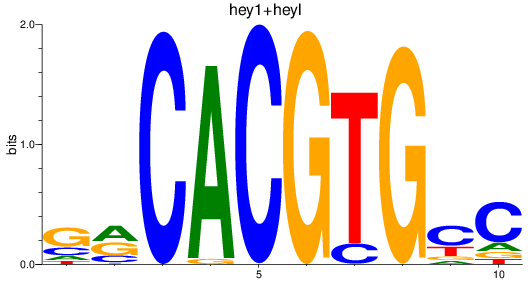
Results for hey2_hey1+heyl
Z-value: 0.66
Transcription factors associated with hey2_hey1+heyl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hey2
|
ENSDARG00000013441 | hes-related family bHLH transcription factor with YRPW motif 2 |
|
heyl
|
ENSDARG00000055798 | hes related family bHLH transcription factor with YRPW motif like |
|
hey1
|
ENSDARG00000070538 | hes-related family bHLH transcription factor with YRPW motif 1 |
|
heyl
|
ENSDARG00000112770 | hes related family bHLH transcription factor with YRPW motif like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| heyl | dr11_v1_chr19_+_32553874_32553874 | -0.32 | 2.0e-01 | Click! |
| hey1 | dr11_v1_chr19_-_31802296_31802296 | -0.27 | 2.8e-01 | Click! |
| hey2 | dr11_v1_chr20_-_39596338_39596338 | 0.10 | 7.0e-01 | Click! |
Activity profile of hey2_hey1+heyl motif
Sorted Z-values of hey2_hey1+heyl motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_68807170 | 1.12 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr7_+_38750871 | 0.86 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr17_+_24318753 | 0.83 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr1_-_41192059 | 0.68 |
ENSDART00000084665
ENSDART00000135369 |
dok7
|
docking protein 7 |
| chr2_-_49860723 | 0.64 |
ENSDART00000083690
|
blvra
|
biliverdin reductase A |
| chr7_+_31319876 | 0.63 |
ENSDART00000187611
|
fam189a1
|
family with sequence similarity 189, member A1 |
| chr7_+_38717624 | 0.60 |
ENSDART00000132522
|
syt13
|
synaptotagmin XIII |
| chr1_-_13989643 | 0.59 |
ENSDART00000191046
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr14_+_22076596 | 0.58 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr10_+_2684958 | 0.58 |
ENSDART00000112019
|
setd9
|
SET domain containing 9 |
| chr5_-_32338866 | 0.57 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_+_29163762 | 0.56 |
ENSDART00000173762
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr21_-_36948 | 0.55 |
ENSDART00000181230
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr1_+_31573225 | 0.54 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr23_+_2421689 | 0.54 |
ENSDART00000180200
|
tcp1
|
t-complex 1 |
| chr2_-_42375275 | 0.54 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr4_-_16824231 | 0.53 |
ENSDART00000014007
|
gys2
|
glycogen synthase 2 |
| chr20_-_14665002 | 0.53 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr21_+_31150438 | 0.52 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr6_+_153146 | 0.52 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr4_-_16824556 | 0.50 |
ENSDART00000165289
ENSDART00000185839 |
gys2
|
glycogen synthase 2 |
| chr18_+_14693682 | 0.49 |
ENSDART00000132249
|
uri1
|
URI1, prefoldin-like chaperone |
| chr11_+_3959495 | 0.46 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr8_-_21372446 | 0.46 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr9_-_12888082 | 0.46 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr21_+_31150773 | 0.45 |
ENSDART00000126205
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr3_+_1150348 | 0.45 |
ENSDART00000148524
|
nol12
|
nucleolar protein 12 |
| chr8_+_26059677 | 0.45 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr7_+_32021982 | 0.44 |
ENSDART00000173848
|
mettl15
|
methyltransferase like 15 |
| chr25_-_35113891 | 0.43 |
ENSDART00000190724
|
zgc:165555
|
zgc:165555 |
| chr7_+_32021669 | 0.43 |
ENSDART00000173976
|
mettl15
|
methyltransferase like 15 |
| chr19_-_35361556 | 0.43 |
ENSDART00000012167
|
ndufs5
|
NADH dehydrogenase (ubiquinone) Fe-S protein 5 |
| chr12_-_33359654 | 0.42 |
ENSDART00000001907
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr21_+_39673157 | 0.40 |
ENSDART00000100240
|
mrm3b
|
mitochondrial rRNA methyltransferase 3b |
| chr23_+_2728095 | 0.40 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr12_+_19384615 | 0.40 |
ENSDART00000078266
|
rsl1d1
|
ribosomal L1 domain containing 1 |
| chr16_-_40459104 | 0.39 |
ENSDART00000032389
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr23_+_2421313 | 0.39 |
ENSDART00000126038
|
tcp1
|
t-complex 1 |
| chr5_+_43470544 | 0.39 |
ENSDART00000111587
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr18_+_5875268 | 0.39 |
ENSDART00000177784
ENSDART00000122009 |
wdr59
|
WD repeat domain 59 |
| chr19_+_7152966 | 0.38 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr15_-_17099560 | 0.37 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr14_-_26704829 | 0.37 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr21_+_1647990 | 0.36 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr10_+_29770120 | 0.36 |
ENSDART00000100032
ENSDART00000193205 |
hyou1
|
hypoxia up-regulated 1 |
| chr23_-_19686791 | 0.36 |
ENSDART00000161973
|
zgc:193598
|
zgc:193598 |
| chr4_-_178510 | 0.35 |
ENSDART00000169805
|
eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr9_-_11587070 | 0.35 |
ENSDART00000030995
|
umps
|
uridine monophosphate synthetase |
| chr25_-_36367523 | 0.34 |
ENSDART00000181344
|
CR354435.4
|
|
| chr1_+_17695426 | 0.34 |
ENSDART00000103236
|
ankrd37
|
ankyrin repeat domain 37 |
| chr24_+_32668675 | 0.34 |
ENSDART00000156638
ENSDART00000155973 |
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr9_-_3149896 | 0.34 |
ENSDART00000020861
|
pdk1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr17_+_53156530 | 0.34 |
ENSDART00000126277
ENSDART00000156774 |
dph6
|
diphthamine biosynthesis 6 |
| chr15_-_44077937 | 0.33 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr16_-_48400639 | 0.32 |
ENSDART00000159372
|
eif3ha
|
eukaryotic translation initiation factor 3, subunit H, a |
| chr6_-_28222592 | 0.32 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr3_+_53773256 | 0.32 |
ENSDART00000170461
|
col5a3a
|
collagen, type V, alpha 3a |
| chr25_-_8625601 | 0.32 |
ENSDART00000155280
|
GDPGP1
|
zgc:153343 |
| chr5_+_27488975 | 0.32 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr20_-_31496679 | 0.31 |
ENSDART00000153437
ENSDART00000153193 |
sash1a
|
SAM and SH3 domain containing 1a |
| chr13_+_41917606 | 0.31 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr24_+_31459715 | 0.31 |
ENSDART00000181102
ENSDART00000189950 ENSDART00000192321 ENSDART00000126380 |
cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr7_+_74134010 | 0.30 |
ENSDART00000164874
|
cldnd1a
|
claudin domain containing 1a |
| chr20_+_50115335 | 0.30 |
ENSDART00000031139
|
slc24a4b
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b |
| chr3_+_23029934 | 0.30 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr14_+_14836468 | 0.30 |
ENSDART00000166728
|
si:dkey-102m7.3
|
si:dkey-102m7.3 |
| chr7_-_41812015 | 0.30 |
ENSDART00000174058
|
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr7_-_30174882 | 0.30 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr7_-_41812355 | 0.29 |
ENSDART00000016105
|
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr24_-_38644937 | 0.29 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr18_-_5875433 | 0.29 |
ENSDART00000151727
|
nob1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr19_-_13933204 | 0.29 |
ENSDART00000158059
|
trnau1apa
|
tRNA selenocysteine 1 associated protein 1a |
| chr18_-_5103931 | 0.28 |
ENSDART00000188091
|
pdcd10a
|
programmed cell death 10a |
| chr6_-_39919982 | 0.28 |
ENSDART00000065091
ENSDART00000064903 |
sumf1
|
sulfatase modifying factor 1 |
| chr24_+_42074143 | 0.28 |
ENSDART00000170514
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr9_+_12887491 | 0.28 |
ENSDART00000102386
|
si:ch211-167j6.4
|
si:ch211-167j6.4 |
| chr4_-_5912951 | 0.28 |
ENSDART00000169439
|
arl1
|
ADP-ribosylation factor-like 1 |
| chr20_-_27733683 | 0.28 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr16_+_19029297 | 0.27 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr25_+_37268900 | 0.27 |
ENSDART00000156737
|
si:dkey-234i14.6
|
si:dkey-234i14.6 |
| chr1_-_26676391 | 0.27 |
ENSDART00000152492
|
trmo
|
tRNA methyltransferase O |
| chr1_-_26675969 | 0.27 |
ENSDART00000054184
|
trmo
|
tRNA methyltransferase O |
| chr18_+_6126506 | 0.27 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr2_+_25839650 | 0.26 |
ENSDART00000134077
ENSDART00000140804 |
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr5_-_26247671 | 0.26 |
ENSDART00000145187
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr5_-_26247215 | 0.26 |
ENSDART00000136806
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr25_-_6223567 | 0.26 |
ENSDART00000067512
|
psma4
|
proteasome subunit alpha 4 |
| chr19_-_34979837 | 0.26 |
ENSDART00000044838
|
ndrg1a
|
N-myc downstream regulated 1a |
| chr5_-_26247973 | 0.25 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr1_+_30946231 | 0.25 |
ENSDART00000022841
|
metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr3_+_28939759 | 0.25 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr12_-_9516981 | 0.25 |
ENSDART00000106285
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| chr7_+_66048102 | 0.25 |
ENSDART00000104523
|
arntl1b
|
aryl hydrocarbon receptor nuclear translocator-like 1b |
| chr2_+_25839940 | 0.24 |
ENSDART00000139927
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr20_+_5065268 | 0.24 |
ENSDART00000053883
|
si:dkey-60a16.1
|
si:dkey-60a16.1 |
| chr22_+_17261801 | 0.24 |
ENSDART00000192978
ENSDART00000193187 ENSDART00000179953 ENSDART00000134798 |
tdrd5
|
tudor domain containing 5 |
| chr16_+_17252487 | 0.24 |
ENSDART00000063572
|
gtf3c6
|
general transcription factor IIIC, polypeptide 6, alpha |
| chr9_+_3283608 | 0.24 |
ENSDART00000192275
|
hat1
|
histone acetyltransferase 1 |
| chr20_-_25626693 | 0.24 |
ENSDART00000132247
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr14_-_52480661 | 0.23 |
ENSDART00000158353
|
exosc3
|
exosome component 3 |
| chr20_-_25626198 | 0.23 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr14_+_24935131 | 0.23 |
ENSDART00000170871
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr20_-_25626428 | 0.23 |
ENSDART00000136475
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr13_-_280652 | 0.23 |
ENSDART00000193627
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr8_-_18582922 | 0.23 |
ENSDART00000123917
|
tmem47
|
transmembrane protein 47 |
| chr9_-_10804796 | 0.23 |
ENSDART00000134911
|
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr18_-_11729 | 0.22 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr2_+_25840463 | 0.22 |
ENSDART00000125178
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr7_-_13362590 | 0.22 |
ENSDART00000091616
|
sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr7_-_14535862 | 0.22 |
ENSDART00000112980
ENSDART00000135446 |
mrps11
|
mitochondrial ribosomal protein S11 |
| chr25_+_25123385 | 0.22 |
ENSDART00000163892
|
ldha
|
lactate dehydrogenase A4 |
| chr9_-_403767 | 0.22 |
ENSDART00000167743
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr2_+_8780443 | 0.22 |
ENSDART00000137768
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr2_-_11504347 | 0.22 |
ENSDART00000019392
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr7_+_14536083 | 0.21 |
ENSDART00000027331
|
mrpl46
|
mitochondrial ribosomal protein L46 |
| chr9_-_10805231 | 0.21 |
ENSDART00000193913
ENSDART00000078348 |
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr22_-_94352 | 0.21 |
ENSDART00000184883
|
ndufaf3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr8_+_39760258 | 0.21 |
ENSDART00000037914
|
cox6a1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr22_-_38035084 | 0.21 |
ENSDART00000145029
|
pex6
|
peroxisomal biogenesis factor 6 |
| chr8_-_26961779 | 0.20 |
ENSDART00000099214
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr20_-_29683754 | 0.20 |
ENSDART00000130599
ENSDART00000015928 ENSDART00000131219 |
si:ch211-195d17.2
|
si:ch211-195d17.2 |
| chr18_+_5850837 | 0.20 |
ENSDART00000013150
|
cog8
|
component of oligomeric golgi complex 8 |
| chr8_-_38317914 | 0.20 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr15_+_28410664 | 0.20 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr12_-_49168398 | 0.20 |
ENSDART00000186608
|
FO704624.1
|
|
| chr11_+_25485774 | 0.20 |
ENSDART00000026249
|
gnl3l
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr6_-_57476465 | 0.20 |
ENSDART00000128065
|
ddx27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr20_-_165818 | 0.20 |
ENSDART00000123860
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr23_+_25292147 | 0.19 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr22_-_38034852 | 0.19 |
ENSDART00000104613
|
pex6
|
peroxisomal biogenesis factor 6 |
| chr22_-_94519 | 0.19 |
ENSDART00000161432
|
ndufaf3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr21_-_2322102 | 0.19 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr13_-_280827 | 0.19 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr2_-_32513538 | 0.19 |
ENSDART00000056640
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr22_-_20695237 | 0.19 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr24_-_20208474 | 0.19 |
ENSDART00000139329
|
cry-dash
|
cryptochrome DASH |
| chr4_-_5913338 | 0.18 |
ENSDART00000183590
|
arl1
|
ADP-ribosylation factor-like 1 |
| chr16_+_22865942 | 0.18 |
ENSDART00000103235
ENSDART00000143957 |
flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr10_+_158590 | 0.18 |
ENSDART00000081982
|
KCNJ15
|
potassium voltage-gated channel subfamily J member 15 |
| chr12_-_33359052 | 0.18 |
ENSDART00000135943
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr7_+_22801465 | 0.18 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr17_-_48705993 | 0.18 |
ENSDART00000030934
|
kcnk5a
|
potassium channel, subfamily K, member 5a |
| chr21_+_40498628 | 0.18 |
ENSDART00000163454
|
coro6
|
coronin 6 |
| chr24_-_42090635 | 0.18 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr25_+_16945348 | 0.18 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr5_+_4366431 | 0.18 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr20_-_32405440 | 0.17 |
ENSDART00000062978
ENSDART00000153411 |
afg1lb
|
AFG1 like ATPase b |
| chr10_+_10972795 | 0.17 |
ENSDART00000127331
|
cdc37l1
|
cell division cycle 37-like 1 |
| chr7_-_41512999 | 0.17 |
ENSDART00000173577
|
si:dkey-10f23.2
|
si:dkey-10f23.2 |
| chr22_-_18779232 | 0.17 |
ENSDART00000186726
|
atp5f1d
|
ATP synthase F1 subunit delta |
| chr12_-_30359031 | 0.17 |
ENSDART00000192628
|
tdrd1
|
tudor domain containing 1 |
| chr10_+_5268054 | 0.17 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr3_-_62380146 | 0.17 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr18_+_38191346 | 0.16 |
ENSDART00000052703
|
nucb2b
|
nucleobindin 2b |
| chr20_+_25626479 | 0.16 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr20_-_13140309 | 0.16 |
ENSDART00000020703
ENSDART00000188594 |
ints7
|
integrator complex subunit 7 |
| chr9_+_42066030 | 0.16 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr20_+_38458084 | 0.16 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr18_+_40993369 | 0.16 |
ENSDART00000141162
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr12_+_33975065 | 0.16 |
ENSDART00000036649
|
sfxn2
|
sideroflexin 2 |
| chr19_-_32518556 | 0.16 |
ENSDART00000103410
|
zbtb8b
|
zinc finger and BTB domain containing 8B |
| chr25_-_35101396 | 0.16 |
ENSDART00000138865
|
zgc:162611
|
zgc:162611 |
| chr13_-_31370184 | 0.16 |
ENSDART00000034829
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr25_-_35101673 | 0.16 |
ENSDART00000140864
|
zgc:162611
|
zgc:162611 |
| chr11_-_44194132 | 0.16 |
ENSDART00000182954
ENSDART00000111271 |
CABZ01080074.1
|
|
| chr9_-_14137295 | 0.16 |
ENSDART00000127640
ENSDART00000189018 ENSDART00000188985 |
ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr18_+_40993196 | 0.16 |
ENSDART00000115111
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr19_-_5103313 | 0.16 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr7_+_40148492 | 0.15 |
ENSDART00000110789
|
esyt2b
|
extended synaptotagmin-like protein 2b |
| chr20_-_25486384 | 0.15 |
ENSDART00000141340
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr10_+_41159241 | 0.15 |
ENSDART00000141657
|
anxa4
|
annexin A4 |
| chr13_+_35925490 | 0.15 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr20_+_32406011 | 0.15 |
ENSDART00000018640
ENSDART00000137910 |
snx3
|
sorting nexin 3 |
| chr18_+_5490668 | 0.15 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr9_-_12594759 | 0.15 |
ENSDART00000021266
|
tra2b
|
transformer 2 beta homolog |
| chr5_-_33959868 | 0.15 |
ENSDART00000143652
|
zgc:63972
|
zgc:63972 |
| chr1_-_19648227 | 0.15 |
ENSDART00000054574
|
polr1e
|
polymerase (RNA) I polypeptide E |
| chr3_+_54887782 | 0.14 |
ENSDART00000018205
|
aanat2
|
arylalkylamine N-acetyltransferase 2 |
| chr1_-_157563 | 0.14 |
ENSDART00000018741
|
pcid2
|
PCI domain containing 2 |
| chr14_-_498979 | 0.14 |
ENSDART00000171976
|
spry1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr8_-_39977026 | 0.14 |
ENSDART00000141707
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr7_-_6340998 | 0.14 |
ENSDART00000172884
|
zgc:165555
|
zgc:165555 |
| chr19_+_32822269 | 0.14 |
ENSDART00000052097
ENSDART00000052096 ENSDART00000188012 |
rida
|
reactive intermediate imine deaminase A homolog |
| chr15_+_6652396 | 0.14 |
ENSDART00000192813
ENSDART00000157678 |
nop53
|
NOP53 ribosome biogenesis factor |
| chr5_-_13167097 | 0.14 |
ENSDART00000149700
ENSDART00000030213 |
mapk1
|
mitogen-activated protein kinase 1 |
| chr8_+_23213320 | 0.14 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr11_+_45299447 | 0.14 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr9_-_35557397 | 0.14 |
ENSDART00000100681
|
ncam2
|
neural cell adhesion molecule 2 |
| chr17_-_24879003 | 0.13 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr7_-_16034324 | 0.13 |
ENSDART00000002498
ENSDART00000162962 |
elp4
|
elongator acetyltransferase complex subunit 4 |
| chr7_+_16033923 | 0.13 |
ENSDART00000161669
ENSDART00000114062 |
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr17_+_52612866 | 0.13 |
ENSDART00000182828
ENSDART00000191156 ENSDART00000188814 ENSDART00000109891 |
angel1
|
angel homolog 1 (Drosophila) |
| chr11_+_25328199 | 0.13 |
ENSDART00000141478
ENSDART00000112209 |
fam83d
|
family with sequence similarity 83, member D |
| chr8_+_21159122 | 0.13 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr24_+_42129775 | 0.13 |
ENSDART00000181576
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr9_+_907459 | 0.13 |
ENSDART00000034850
ENSDART00000144114 |
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr8_+_23738122 | 0.13 |
ENSDART00000062983
|
rpl10a
|
ribosomal protein L10a |
| chr11_-_438294 | 0.13 |
ENSDART00000040812
|
nuf2
|
NUF2, NDC80 kinetochore complex component, homolog |
| chr7_+_67467702 | 0.13 |
ENSDART00000168460
ENSDART00000170322 |
rpl13
|
ribosomal protein L13 |
| chr19_-_791016 | 0.13 |
ENSDART00000037515
|
msto1
|
misato 1, mitochondrial distribution and morphology regulator |
Network of associatons between targets according to the STRING database.
First level regulatory network of hey2_hey1+heyl
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.4 | GO:1902102 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 0.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.7 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.3 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 0.9 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.8 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.2 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.5 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 0.1 | 0.6 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.6 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 0.8 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.3 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.2 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 0.2 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 0.2 | GO:0072388 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.1 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.2 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.4 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.8 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.1 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.0 | 0.2 | GO:0032206 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.4 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.2 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.2 | GO:0097032 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 1.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.9 | GO:0009749 | response to glucose(GO:0009749) |
| 0.0 | 0.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.0 | 0.2 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0019478 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.0 | 0.3 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.0 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.6 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.4 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.5 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.7 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0019079 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.0 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.1 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.0 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.0 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.4 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.4 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.0 | GO:0060585 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 1.4 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.3 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.1 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 1.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.3 | 1.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 0.7 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 1.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.5 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.5 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 0.2 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.1 | 0.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.3 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.2 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 1.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.5 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.8 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.7 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |