Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
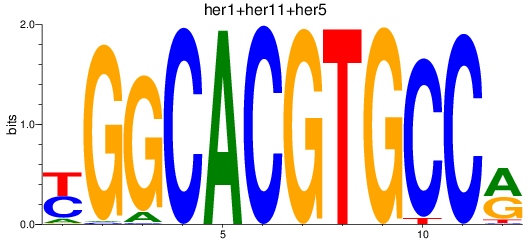
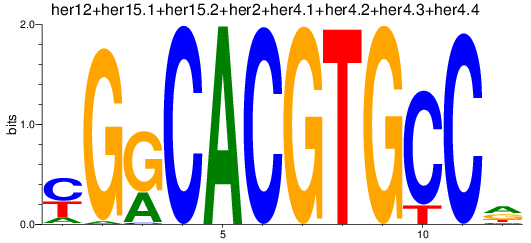
Results for her1+her11+her5_her12+her15.1+her15.2+her2+her4.1+her4.2+her4.3+her4.4
Z-value: 0.17
Transcription factors associated with her1+her11+her5_her12+her15.1+her15.2+her2+her4.1+her4.2+her4.3+her4.4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
her11
|
ENSDARG00000002707 | hairy-related 11 |
|
her5
|
ENSDARG00000008796 | hairy-related 5 |
|
her1
|
ENSDARG00000014722 | hairy-related 1 |
|
her4.4
|
ENSDARG00000009822 | hairy-related 4, tandem duplicate 4 |
|
her12
|
ENSDARG00000032963 | hairy-related 12 |
|
her2
|
ENSDARG00000038205 | hairy-related 2 |
|
her15.2
|
ENSDARG00000054560 | hairy and enhancer of split-related 15, tandem duplicate 2 |
|
her15.1
|
ENSDARG00000054562 | hairy and enhancer of split-related 15, tandem duplicate 1 |
|
her4.2
|
ENSDARG00000056729 | hairy-related 4, tandem duplicate 2 |
|
her4.1
|
ENSDARG00000056732 | hairy-related 4, tandem duplicate 1 |
|
her4.3
|
ENSDARG00000070770 | hairy-related 4, tandem duplicate 3 |
|
her4.2
|
ENSDARG00000094426 | hairy-related 4, tandem duplicate 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| her2 | dr11_v1_chr11_-_41966854_41966854 | 0.60 | 7.9e-03 | Click! |
| her1 | dr11_v1_chr5_-_68795063_68795063 | -0.55 | 1.7e-02 | Click! |
| her4.3 | dr11_v1_chr23_+_21459263_21459263 | 0.46 | 5.2e-02 | Click! |
| her4.4 | dr11_v1_chr23_-_21463788_21463788 | 0.39 | 1.1e-01 | Click! |
| her11 | dr11_v1_chr14_-_30540967_30540967 | 0.24 | 3.3e-01 | Click! |
| her4.2 | dr11_v1_chr23_+_21455152_21455154 | 0.24 | 3.4e-01 | Click! |
| her12 | dr11_v1_chr23_-_21446985_21446985 | 0.20 | 4.3e-01 | Click! |
| her15.2 | dr11_v1_chr11_-_41996957_41996957 | 0.15 | 5.4e-01 | Click! |
| her5 | dr11_v1_chr14_+_30543008_30543008 | -0.12 | 6.5e-01 | Click! |
| her4.1 | dr11_v1_chr23_-_21453614_21453614 | 0.09 | 7.2e-01 | Click! |
| her15.1 | dr11_v1_chr11_+_41981959_41981959 | -0.03 | 9.0e-01 | Click! |
Activity profile of her1+her11+her5_her12+her15.1+her15.2+her2+her4.1+her4.2+her4.3+her4.4 motif
Sorted Z-values of her1+her11+her5_her12+her15.1+her15.2+her2+her4.1+her4.2+her4.3+her4.4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_62723233 | 0.78 |
ENSDART00000183718
|
nanos2
|
nanos homolog 2 |
| chr16_-_45069882 | 0.77 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr14_+_46313135 | 0.49 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr7_-_40993456 | 0.35 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr10_-_27566481 | 0.31 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr2_+_29976419 | 0.29 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr14_+_46313396 | 0.20 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr6_+_39184236 | 0.16 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr11_-_3959477 | 0.13 |
ENSDART00000045971
|
pbrm1
|
polybromo 1 |
| chr9_+_3283608 | 0.12 |
ENSDART00000192275
|
hat1
|
histone acetyltransferase 1 |
| chr11_-_3959889 | 0.11 |
ENSDART00000159683
|
pbrm1
|
polybromo 1 |
| chr19_+_31183495 | 0.11 |
ENSDART00000088618
|
meox2b
|
mesenchyme homeobox 2b |
| chr5_-_46329880 | 0.11 |
ENSDART00000156577
|
si:ch211-130m23.5
|
si:ch211-130m23.5 |
| chr15_-_14469704 | 0.11 |
ENSDART00000185077
|
numbl
|
numb homolog (Drosophila)-like |
| chr8_+_39760258 | 0.11 |
ENSDART00000037914
|
cox6a1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr19_+_42470396 | 0.10 |
ENSDART00000191679
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr4_-_8093753 | 0.10 |
ENSDART00000133434
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr23_+_21459263 | 0.09 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr9_-_44295071 | 0.09 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr14_+_14836468 | 0.08 |
ENSDART00000166728
|
si:dkey-102m7.3
|
si:dkey-102m7.3 |
| chr11_+_3959495 | 0.08 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr3_+_32425202 | 0.07 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr6_+_49021703 | 0.07 |
ENSDART00000149394
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr4_-_44611273 | 0.06 |
ENSDART00000156793
|
znf1115
|
zinc finger protein 1115 |
| chr19_+_42142381 | 0.05 |
ENSDART00000129915
|
kcnq4
|
potassium voltage-gated channel, KQT-like subfamily, member 4 |
| chr19_-_35596207 | 0.05 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr9_+_45789887 | 0.05 |
ENSDART00000135202
|
si:dkey-34f9.3
|
si:dkey-34f9.3 |
| chr23_-_21463788 | 0.04 |
ENSDART00000079265
|
her4.4
|
hairy-related 4, tandem duplicate 4 |
| chr19_+_42469058 | 0.04 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr8_-_39760427 | 0.04 |
ENSDART00000184336
ENSDART00000186500 |
si:ch211-170d8.8
|
si:ch211-170d8.8 |
| chr20_-_39344116 | 0.04 |
ENSDART00000037282
|
ccdc25
|
coiled-coil domain containing 25 |
| chr14_-_30387894 | 0.03 |
ENSDART00000176136
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr2_-_32386791 | 0.03 |
ENSDART00000056634
|
ubtfl
|
upstream binding transcription factor, like |
| chr8_+_29593986 | 0.03 |
ENSDART00000077642
|
atoh1a
|
atonal bHLH transcription factor 1a |
| chr1_+_5576151 | 0.02 |
ENSDART00000109756
|
cpo
|
carboxypeptidase O |
| chr11_+_29671661 | 0.02 |
ENSDART00000024318
ENSDART00000165024 |
rnf207a
|
ring finger protein 207a |
| chr12_+_30046320 | 0.02 |
ENSDART00000179904
ENSDART00000153394 |
ablim1b
|
actin binding LIM protein 1b |
| chr20_+_19212962 | 0.00 |
ENSDART00000063706
|
fndc4a
|
fibronectin type III domain containing 4a |
Network of associatons between targets according to the STRING database.
First level regulatory network of her1+her11+her5_her12+her15.1+her15.2+her2+her4.1+her4.2+her4.3+her4.4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.4 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.8 | GO:0006096 | glycolytic process(GO:0006096) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016586 | RSC complex(GO:0016586) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |