Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
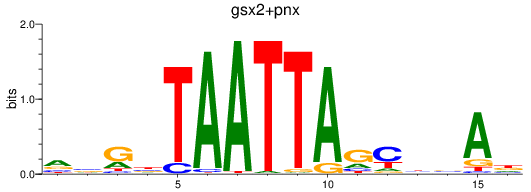
Results for gsx2+pnx
Z-value: 1.35
Transcription factors associated with gsx2+pnx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pnx
|
ENSDARG00000025899 | posterior neuron-specific homeobox |
|
gsx2
|
ENSDARG00000043322 | GS homeobox 2 |
|
gsx2
|
ENSDARG00000116417 | GS homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pnx | dr11_v1_chr10_+_2799285_2799285 | 0.97 | 8.6e-11 | Click! |
| gsx2 | dr11_v1_chr20_-_22484621_22484621 | 0.64 | 4.3e-03 | Click! |
Activity profile of gsx2+pnx motif
Sorted Z-values of gsx2+pnx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_33865312 | 6.56 |
ENSDART00000166141
|
tekt1
|
tektin 1 |
| chr7_-_1504382 | 5.25 |
ENSDART00000172770
|
si:zfos-405g10.4
|
si:zfos-405g10.4 |
| chr10_-_17083180 | 4.61 |
ENSDART00000170083
|
fam166b
|
family with sequence similarity 166, member B |
| chr11_-_40728380 | 4.57 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr6_-_40352215 | 4.39 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr2_-_2957970 | 4.35 |
ENSDART00000162505
|
si:ch1073-82l19.1
|
si:ch1073-82l19.1 |
| chr4_+_306036 | 4.17 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr6_+_40354424 | 4.00 |
ENSDART00000047416
|
slc4a8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr5_+_42308955 | 3.58 |
ENSDART00000136683
|
pimr201
|
Pim proto-oncogene, serine/threonine kinase, related 201 |
| chr18_+_1154189 | 3.35 |
ENSDART00000135090
|
si:ch1073-75f15.2
|
si:ch1073-75f15.2 |
| chr20_-_14925281 | 3.31 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr11_-_472547 | 3.19 |
ENSDART00000005923
|
zgc:77375
|
zgc:77375 |
| chr1_-_17715493 | 3.18 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr20_-_1378514 | 3.03 |
ENSDART00000181830
|
scara5
|
scavenger receptor class A, member 5 (putative) |
| chr19_+_43978814 | 2.94 |
ENSDART00000102314
|
nkd3
|
naked cuticle homolog 3 |
| chr21_+_43404945 | 2.90 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr20_-_14924858 | 2.89 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr5_-_13564961 | 2.85 |
ENSDART00000146827
|
si:ch211-230g14.3
|
si:ch211-230g14.3 |
| chr1_-_59287410 | 2.84 |
ENSDART00000158011
ENSDART00000170580 |
col5a3b
|
collagen, type V, alpha 3b |
| chr21_-_29991098 | 2.75 |
ENSDART00000155772
|
si:ch73-190f9.4
|
si:ch73-190f9.4 |
| chr8_-_21372446 | 2.71 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr8_-_24502997 | 2.68 |
ENSDART00000184801
ENSDART00000179794 |
BX296549.1
|
|
| chr2_-_1569250 | 2.65 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr6_+_3280939 | 2.62 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr6_-_7720332 | 2.56 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr16_-_13818061 | 2.54 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr1_+_50921266 | 2.30 |
ENSDART00000006538
|
otx2a
|
orthodenticle homeobox 2a |
| chr23_+_19813677 | 2.29 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr23_+_45966436 | 2.28 |
ENSDART00000172160
|
CABZ01069338.1
|
|
| chr5_-_65969959 | 2.18 |
ENSDART00000170677
|
ttc16
|
tetratricopeptide repeat domain 16 |
| chr18_+_3332999 | 2.16 |
ENSDART00000160857
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr25_+_31868268 | 2.11 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr7_-_24046999 | 2.11 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr18_-_36909773 | 2.11 |
ENSDART00000141694
|
si:ch211-160d20.5
|
si:ch211-160d20.5 |
| chr13_-_6218248 | 2.02 |
ENSDART00000159052
|
si:zfos-1056e6.1
|
si:zfos-1056e6.1 |
| chr3_-_39180048 | 1.98 |
ENSDART00000049720
|
cdk21
|
cyclin-dependent kinase 21 |
| chr3_-_23643751 | 1.93 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr10_+_35257651 | 1.93 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr13_+_8677166 | 1.92 |
ENSDART00000181016
ENSDART00000135028 |
prop1
|
PROP paired-like homeobox 1 |
| chr5_+_69747417 | 1.87 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr5_-_19394440 | 1.85 |
ENSDART00000163771
|
foxn4
|
forkhead box N4 |
| chr20_+_535458 | 1.85 |
ENSDART00000152453
|
dse
|
dermatan sulfate epimerase |
| chr21_-_5205617 | 1.85 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr2_-_43124747 | 1.82 |
ENSDART00000188883
|
lrrc6
|
leucine rich repeat containing 6 |
| chr15_-_36533322 | 1.76 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr17_+_3379673 | 1.76 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr24_-_8777781 | 1.75 |
ENSDART00000082362
ENSDART00000177400 |
mak
|
male germ cell-associated kinase |
| chr25_+_5035343 | 1.70 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr15_-_4528326 | 1.68 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr1_-_58963395 | 1.66 |
ENSDART00000130415
|
CABZ01115881.1
|
|
| chr15_-_4969525 | 1.60 |
ENSDART00000157223
|
lipt2
|
lipoyl(octanoyl) transferase 2 |
| chr3_+_62356578 | 1.59 |
ENSDART00000157030
|
iqck
|
IQ motif containing K |
| chr21_-_22827548 | 1.56 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr3_+_30500968 | 1.54 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr5_+_19343880 | 1.51 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr17_+_43908428 | 1.51 |
ENSDART00000180332
|
msh4
|
mutS homolog 4 |
| chr20_-_46362606 | 1.49 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr7_-_48667056 | 1.47 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr13_+_30903816 | 1.47 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr14_-_36320506 | 1.47 |
ENSDART00000074639
|
egf
|
epidermal growth factor |
| chr4_+_72723304 | 1.45 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr15_+_15856178 | 1.41 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr11_+_26476153 | 1.39 |
ENSDART00000103507
|
unm_sa1614
|
un-named sa1614 |
| chr19_+_3826782 | 1.39 |
ENSDART00000169222
|
oscp1a
|
organic solute carrier partner 1a |
| chr21_+_6751405 | 1.39 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr3_-_23407720 | 1.37 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr18_-_48547564 | 1.37 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr21_-_45077429 | 1.34 |
ENSDART00000187268
ENSDART00000191003 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr24_-_3783497 | 1.34 |
ENSDART00000158354
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr9_-_950004 | 1.33 |
ENSDART00000168917
|
SCTR
|
secretin receptor |
| chr2_-_43123949 | 1.32 |
ENSDART00000075347
ENSDART00000132346 |
lrrc6
|
leucine rich repeat containing 6 |
| chr10_-_5847655 | 1.31 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr12_+_20352400 | 1.30 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr3_+_18398876 | 1.28 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr13_-_42400647 | 1.28 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr10_-_26744131 | 1.26 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr21_+_25198637 | 1.25 |
ENSDART00000164972
|
si:dkey-183i3.6
|
si:dkey-183i3.6 |
| chr21_-_45076778 | 1.24 |
ENSDART00000181525
|
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr1_+_7517454 | 1.24 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr10_+_26645953 | 1.23 |
ENSDART00000131482
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr16_+_6021908 | 1.22 |
ENSDART00000163786
|
BX511115.1
|
|
| chr15_+_1796313 | 1.22 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr24_-_38657683 | 1.21 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr15_-_21877726 | 1.21 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr20_-_1176631 | 1.20 |
ENSDART00000152255
|
gabrr2b
|
gamma-aminobutyric acid (GABA) A receptor, rho 2b |
| chr21_+_6751760 | 1.19 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr12_+_30265802 | 1.18 |
ENSDART00000182412
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr19_-_7495006 | 1.18 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr6_+_9677420 | 1.16 |
ENSDART00000064995
|
sumo1
|
small ubiquitin-like modifier 1 |
| chr16_+_41826584 | 1.16 |
ENSDART00000147523
|
si:dkey-199f5.6
|
si:dkey-199f5.6 |
| chr10_+_6884627 | 1.14 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr1_-_19502322 | 1.14 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr11_+_7264457 | 1.13 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr20_-_9095105 | 1.12 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr6_+_6924637 | 1.10 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr4_+_11464255 | 1.10 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr18_-_43884044 | 1.10 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr2_+_33051730 | 1.09 |
ENSDART00000177902
ENSDART00000187903 |
rnf220a
|
ring finger protein 220a |
| chr11_+_33818179 | 1.08 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr18_+_7639401 | 1.08 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr4_+_22480169 | 1.08 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr19_-_31402429 | 1.07 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr6_+_41191482 | 1.05 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr12_-_28363111 | 1.05 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr25_+_31267268 | 1.04 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr18_-_21950751 | 1.04 |
ENSDART00000043452
|
tsnaxip1
|
translin-associated factor X interacting protein 1 |
| chr20_-_48470599 | 1.04 |
ENSDART00000166857
|
CABZ01059120.1
|
|
| chr16_-_42933332 | 1.01 |
ENSDART00000057305
|
thbs3a
|
thrombospondin 3a |
| chr23_+_26979174 | 0.99 |
ENSDART00000018654
|
rnd1b
|
Rho family GTPase 1b |
| chr16_-_51271962 | 0.99 |
ENSDART00000164021
ENSDART00000046420 |
serpinb1l1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 1 |
| chr23_+_20110086 | 0.98 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr15_-_22074315 | 0.97 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr21_+_45841731 | 0.97 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr15_-_35252522 | 0.96 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr11_-_2131280 | 0.95 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr3_+_29445473 | 0.94 |
ENSDART00000020381
|
grap2a
|
GRB2-related adaptor protein 2a |
| chr8_+_20776654 | 0.93 |
ENSDART00000135850
|
nfic
|
nuclear factor I/C |
| chr18_+_14529005 | 0.93 |
ENSDART00000186379
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr24_+_12133814 | 0.93 |
ENSDART00000158562
ENSDART00000159029 ENSDART00000168248 |
lztfl1
|
leucine zipper transcription factor-like 1 |
| chr6_-_40832245 | 0.92 |
ENSDART00000007783
|
eefsec
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr9_-_813511 | 0.90 |
ENSDART00000082368
|
marco
|
macrophage receptor with collagenous structure |
| chr3_+_59784632 | 0.89 |
ENSDART00000084729
|
pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr9_-_53666031 | 0.89 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr1_-_50859053 | 0.89 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr13_-_36663358 | 0.88 |
ENSDART00000085319
|
sos2
|
son of sevenless homolog 2 (Drosophila) |
| chr25_+_20715950 | 0.87 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr10_+_42690374 | 0.87 |
ENSDART00000123496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr18_+_5137241 | 0.86 |
ENSDART00000159601
|
serpini1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr4_+_74929427 | 0.86 |
ENSDART00000174082
|
nup50
|
nucleoporin 50 |
| chr15_+_41027466 | 0.85 |
ENSDART00000075940
|
mtnr1ba
|
melatonin receptor type 1Ba |
| chr19_-_5103313 | 0.85 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr9_-_22834860 | 0.85 |
ENSDART00000146486
|
neb
|
nebulin |
| chr14_-_7137808 | 0.85 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr13_+_22295905 | 0.84 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr23_-_16485190 | 0.84 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr11_+_25129013 | 0.84 |
ENSDART00000132879
|
ndrg3a
|
ndrg family member 3a |
| chr4_+_76575585 | 0.83 |
ENSDART00000131588
|
ms4a17a.11
|
membrane-spanning 4-domains, subfamily A, member 17A.11 |
| chr23_+_40460333 | 0.82 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr20_-_47188966 | 0.81 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr5_+_40485503 | 0.81 |
ENSDART00000051055
|
ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) |
| chr22_-_15578402 | 0.79 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr19_-_5103141 | 0.79 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr6_-_30210378 | 0.77 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr2_+_6963296 | 0.76 |
ENSDART00000147146
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr4_+_58576146 | 0.76 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr3_+_32112004 | 0.75 |
ENSDART00000105272
|
zgc:173593
|
zgc:173593 |
| chr4_+_71989418 | 0.75 |
ENSDART00000170996
|
parp11
|
poly(ADP-ribose) polymerase family member 11 |
| chr20_+_46513651 | 0.75 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr11_-_41853874 | 0.74 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr19_-_29853402 | 0.73 |
ENSDART00000024292
ENSDART00000188508 |
txlna
|
taxilin alpha |
| chr18_+_17827149 | 0.73 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr14_-_40390757 | 0.73 |
ENSDART00000149443
|
pcdh19
|
protocadherin 19 |
| chr12_+_5977777 | 0.72 |
ENSDART00000152302
|
si:ch211-131k2.2
|
si:ch211-131k2.2 |
| chr24_-_31425799 | 0.72 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr8_+_52637507 | 0.72 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr11_+_38280454 | 0.71 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr4_+_47636303 | 0.71 |
ENSDART00000167272
ENSDART00000166961 |
BX324142.1
|
|
| chr18_+_11506561 | 0.71 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr16_-_31754102 | 0.71 |
ENSDART00000185043
|
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr15_+_23799461 | 0.70 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr13_+_35339182 | 0.69 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr8_+_52619365 | 0.69 |
ENSDART00000162953
|
cyr61l2
|
cysteine-rich, angiogenic inducer, 61 like 2 |
| chr14_-_4145594 | 0.69 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr22_-_8388678 | 0.69 |
ENSDART00000184389
|
CABZ01061498.1
|
|
| chr4_-_948776 | 0.69 |
ENSDART00000023483
|
sim1b
|
single-minded family bHLH transcription factor 1b |
| chr4_+_77943184 | 0.67 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr3_-_30625219 | 0.67 |
ENSDART00000151698
|
syt3
|
synaptotagmin III |
| chr15_+_47492838 | 0.65 |
ENSDART00000122372
|
sik3
|
SIK family kinase 3 |
| chr9_-_6372535 | 0.64 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr3_-_60827402 | 0.64 |
ENSDART00000053494
|
anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr23_-_31512496 | 0.63 |
ENSDART00000158755
ENSDART00000143425 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr23_+_44374041 | 0.63 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr21_-_37790727 | 0.63 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr18_-_46258612 | 0.62 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr18_-_2433011 | 0.61 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr17_-_3815693 | 0.61 |
ENSDART00000161207
|
PLCB4
|
phospholipase C beta 4 |
| chr25_+_29160102 | 0.61 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr14_+_11458044 | 0.61 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr5_-_69649044 | 0.60 |
ENSDART00000011192
ENSDART00000170885 |
eif2b1
|
eukaryotic translation initiation factor 2B, subunit 1 alpha |
| chr1_-_14332283 | 0.59 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr19_-_38611814 | 0.58 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr25_-_31953516 | 0.57 |
ENSDART00000110180
|
duox2
|
dual oxidase 2 |
| chr19_+_5543072 | 0.56 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr21_-_26490186 | 0.55 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr24_-_6078222 | 0.55 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr15_+_44366556 | 0.54 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr8_+_29986265 | 0.54 |
ENSDART00000148258
|
ptch1
|
patched 1 |
| chr11_-_15296805 | 0.54 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr22_-_209741 | 0.53 |
ENSDART00000171954
|
ora1
|
olfactory receptor class A related 1 |
| chr5_+_9147497 | 0.53 |
ENSDART00000003273
|
rchy1
|
ring finger and CHY zinc finger domain containing 1 |
| chr5_+_66433287 | 0.52 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr3_-_7546740 | 0.52 |
ENSDART00000128960
|
zmp:0000001003
|
zmp:0000001003 |
| chr4_-_55459037 | 0.52 |
ENSDART00000172184
|
znf569l
|
zinc finger protein 569, like |
| chr18_+_48423973 | 0.51 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr6_-_40884453 | 0.50 |
ENSDART00000017968
ENSDART00000154100 |
sirt4
|
sirtuin 4 |
| chr21_+_18997511 | 0.49 |
ENSDART00000145591
|
rpl17
|
ribosomal protein L17 |
| chr22_-_36530902 | 0.48 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr18_+_23408073 | 0.47 |
ENSDART00000136489
|
mctp2a
|
multiple C2 domains, transmembrane 2a |
| chr20_+_35382482 | 0.47 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr18_-_502722 | 0.47 |
ENSDART00000185757
|
sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gsx2+pnx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.5 | 1.6 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.5 | 6.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.5 | 1.9 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.4 | 3.0 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.4 | 1.6 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 1.2 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.4 | 1.5 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.4 | 1.9 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.3 | 2.6 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.3 | 6.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.3 | 1.9 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 0.9 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.3 | 1.2 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.3 | 1.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.2 | 0.7 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 1.2 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.2 | 0.6 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 1.5 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 1.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 1.0 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.2 | 1.9 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.2 | 4.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.7 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 1.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 1.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.5 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 2.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 1.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 2.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.6 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 1.9 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 1.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.8 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.9 | GO:1903828 | negative regulation of cellular protein localization(GO:1903828) |
| 0.1 | 1.9 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.1 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 1.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 2.6 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 3.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.7 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.3 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 1.1 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 1.1 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 1.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.5 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 5.0 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.1 | 1.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 1.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 0.2 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 0.3 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 0.3 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.1 | 0.8 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 0.3 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.3 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.3 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.7 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 1.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.3 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 1.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.9 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.2 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 2.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 1.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.0 | 2.6 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.7 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.5 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 5.7 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) negative regulation of nuclear division(GO:0051784) |
| 0.0 | 0.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.5 | GO:0046474 | glycerophospholipid biosynthetic process(GO:0046474) |
| 0.0 | 1.4 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.8 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 1.6 | GO:0007163 | establishment or maintenance of cell polarity(GO:0007163) |
| 0.0 | 0.1 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.9 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.7 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.2 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.4 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.9 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.1 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.3 | 6.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.3 | 4.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 1.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 1.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 6.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.9 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 3.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 4.4 | GO:0005930 | axoneme(GO:0005930) |
| 0.1 | 1.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 1.7 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.1 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.2 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 2.8 | GO:0005604 | collagen trimer(GO:0005581) basement membrane(GO:0005604) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.1 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 1.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.9 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 2.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.6 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.9 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.5 | 1.6 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.4 | 1.6 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.4 | 1.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.4 | 1.9 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.4 | 1.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.3 | 4.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 0.7 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.2 | 1.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 1.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 1.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 1.9 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 2.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 1.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 0.5 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 0.9 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 1.0 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.5 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 1.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 2.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.8 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 1.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 1.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 2.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 3.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.7 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 1.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 1.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 1.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0042164 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 1.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.4 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.7 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.6 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 2.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.9 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 1.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.5 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 3.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 6.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 5.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.4 | GO:0019956 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.8 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 3.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 4.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.9 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 2.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 2.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 1.1 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.4 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |