Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
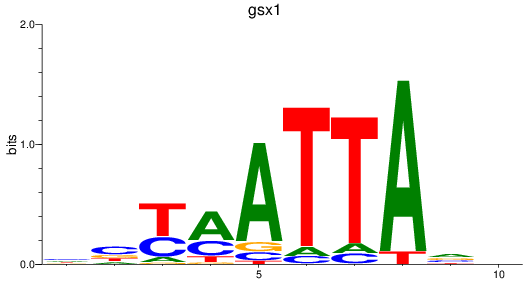
Results for gsx1
Z-value: 0.49
Transcription factors associated with gsx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gsx1
|
ENSDARG00000035735 | GS homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gsx1 | dr11_v1_chr5_-_67911111_67911111 | 0.36 | 1.4e-01 | Click! |
Activity profile of gsx1 motif
Sorted Z-values of gsx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_13818743 | 0.71 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr23_+_20705849 | 0.70 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr20_-_38787341 | 0.51 |
ENSDART00000136771
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr15_-_37104165 | 0.47 |
ENSDART00000165867
|
zmp:0000001114
|
zmp:0000001114 |
| chr16_-_17347727 | 0.46 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr7_+_17816006 | 0.45 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr25_-_5119162 | 0.43 |
ENSDART00000153961
|
shisal1b
|
shisa like 1b |
| chr9_-_40873934 | 0.41 |
ENSDART00000066424
|
pofut2
|
protein O-fucosyltransferase 2 |
| chr8_+_23355484 | 0.41 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chrM_+_9735 | 0.41 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr11_+_10909183 | 0.40 |
ENSDART00000064860
|
rbms1a
|
RNA binding motif, single stranded interacting protein 1a |
| chr7_+_17816470 | 0.40 |
ENSDART00000173807
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr10_+_6884627 | 0.35 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr15_+_34988148 | 0.34 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr24_-_2450597 | 0.33 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr19_+_1688727 | 0.32 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr10_+_6884123 | 0.32 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr20_-_38787047 | 0.31 |
ENSDART00000152913
ENSDART00000153430 |
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr21_+_43404945 | 0.31 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr8_+_17168114 | 0.31 |
ENSDART00000183901
|
cenph
|
centromere protein H |
| chr18_+_15841449 | 0.31 |
ENSDART00000141800
ENSDART00000091349 |
eea1
|
early endosome antigen 1 |
| chr2_-_57076687 | 0.30 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr15_-_43284021 | 0.29 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr7_+_1467863 | 0.29 |
ENSDART00000173433
|
emc4
|
ER membrane protein complex subunit 4 |
| chr19_+_46237665 | 0.29 |
ENSDART00000159391
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr25_-_27722614 | 0.28 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr8_-_50888806 | 0.27 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr21_+_45717930 | 0.26 |
ENSDART00000164315
|
ddx46
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr7_+_756942 | 0.26 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr12_+_28888975 | 0.26 |
ENSDART00000076362
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr22_-_881725 | 0.25 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr14_+_23717165 | 0.25 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr4_+_11723852 | 0.25 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr24_-_11506054 | 0.25 |
ENSDART00000140217
ENSDART00000106310 |
prpf4bb
|
pre-mRNA processing factor 4Bb |
| chr7_-_7764287 | 0.25 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr11_-_2478374 | 0.24 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr21_-_23110841 | 0.24 |
ENSDART00000147896
ENSDART00000003076 ENSDART00000184925 ENSDART00000190386 |
usp28
|
ubiquitin specific peptidase 28 |
| chr16_-_28658341 | 0.24 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr1_+_55239160 | 0.23 |
ENSDART00000152318
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr25_+_28825657 | 0.23 |
ENSDART00000153625
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr14_-_33945692 | 0.23 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr4_+_9177997 | 0.22 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr20_+_34671386 | 0.22 |
ENSDART00000152836
ENSDART00000138226 |
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr23_+_40460333 | 0.22 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr1_-_23157583 | 0.22 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr6_+_4229360 | 0.21 |
ENSDART00000191347
ENSDART00000130642 |
FO082877.1
|
|
| chr3_-_30488063 | 0.21 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr16_-_24605969 | 0.20 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr11_-_42980535 | 0.20 |
ENSDART00000181160
ENSDART00000192064 |
CABZ01069998.1
|
|
| chr17_+_46818521 | 0.20 |
ENSDART00000156022
|
pimr14
|
Pim proto-oncogene, serine/threonine kinase, related 14 |
| chr19_-_31402429 | 0.20 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr3_-_40054615 | 0.19 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr1_+_33668236 | 0.19 |
ENSDART00000122316
ENSDART00000102184 |
arl13b
|
ADP-ribosylation factor-like 13b |
| chr3_-_29941357 | 0.19 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr13_+_48359573 | 0.19 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr20_+_39457598 | 0.19 |
ENSDART00000140931
ENSDART00000156176 |
pimr128
|
Pim proto-oncogene, serine/threonine kinase, related 128 |
| chr21_-_45077429 | 0.18 |
ENSDART00000187268
ENSDART00000191003 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr3_+_45365098 | 0.18 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr2_+_20793982 | 0.18 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr18_+_2228737 | 0.18 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr6_-_2134581 | 0.17 |
ENSDART00000175478
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr8_+_24747865 | 0.17 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr23_+_20563779 | 0.17 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr8_-_25034411 | 0.17 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr19_-_5805923 | 0.17 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr19_-_30510259 | 0.17 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr11_-_35171768 | 0.17 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr1_+_55293424 | 0.16 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr6_+_41191482 | 0.16 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr12_-_18577983 | 0.16 |
ENSDART00000193262
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr15_+_46356879 | 0.15 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr15_+_46357080 | 0.15 |
ENSDART00000155571
ENSDART00000156541 |
wu:fb18f06
|
wu:fb18f06 |
| chr15_+_36309070 | 0.15 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr14_+_7377552 | 0.15 |
ENSDART00000142158
ENSDART00000141471 |
hars
|
histidyl-tRNA synthetase |
| chr21_-_22827548 | 0.15 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr10_+_21867307 | 0.15 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr21_-_20381481 | 0.15 |
ENSDART00000115236
|
atp5mea
|
ATP synthase membrane subunit ea |
| chr9_-_21918963 | 0.15 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr23_-_36303216 | 0.14 |
ENSDART00000188720
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr24_+_5912635 | 0.14 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr14_+_33329761 | 0.14 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr6_-_39024538 | 0.13 |
ENSDART00000165839
|
tns2b
|
tensin 2b |
| chr10_+_8197827 | 0.13 |
ENSDART00000026244
|
mtrex
|
Mtr4 exosome RNA helicase |
| chr10_+_16036246 | 0.13 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr24_-_6078222 | 0.13 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr2_-_55298075 | 0.13 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr19_-_82504 | 0.13 |
ENSDART00000027864
ENSDART00000160560 |
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr6_-_14292307 | 0.12 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr15_-_23376541 | 0.12 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr3_+_46635527 | 0.12 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr3_+_46459540 | 0.12 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr23_+_1033668 | 0.12 |
ENSDART00000143391
|
si:zfos-905g2.1
|
si:zfos-905g2.1 |
| chr2_+_41526904 | 0.12 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr23_-_31913231 | 0.12 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr21_+_28445052 | 0.11 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr12_+_48803098 | 0.11 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr24_+_1023839 | 0.11 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr23_-_31913069 | 0.11 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr18_-_43884044 | 0.11 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr14_-_689841 | 0.11 |
ENSDART00000125969
|
ube2ka
|
ubiquitin-conjugating enzyme E2Ka (UBC1 homolog, yeast) |
| chr2_+_19578446 | 0.11 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr2_+_3986083 | 0.11 |
ENSDART00000188979
|
mkxb
|
mohawk homeobox b |
| chr8_+_11425048 | 0.11 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr13_+_29238850 | 0.11 |
ENSDART00000026000
|
myofl
|
myoferlin like |
| chr15_-_25518084 | 0.10 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr8_-_23612462 | 0.10 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr18_-_2549198 | 0.10 |
ENSDART00000186516
|
CABZ01070631.1
|
|
| chr25_-_7705487 | 0.10 |
ENSDART00000128099
|
prdm11
|
PR domain containing 11 |
| chr4_+_2655358 | 0.10 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr15_+_1796313 | 0.10 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr24_+_19415124 | 0.10 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr9_+_41156818 | 0.10 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr4_+_9400012 | 0.09 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr21_-_17296789 | 0.09 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr15_-_4969525 | 0.09 |
ENSDART00000157223
|
lipt2
|
lipoyl(octanoyl) transferase 2 |
| chr17_-_200316 | 0.09 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr20_+_19512727 | 0.09 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr15_-_1822548 | 0.09 |
ENSDART00000082026
ENSDART00000180230 |
mmp28
|
matrix metallopeptidase 28 |
| chr8_+_21353878 | 0.08 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr20_-_5052786 | 0.08 |
ENSDART00000138818
ENSDART00000181655 ENSDART00000164274 |
arid1b
|
AT rich interactive domain 1B (SWI1-like) |
| chr25_-_37084032 | 0.08 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr7_+_20503344 | 0.08 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr7_+_38509333 | 0.08 |
ENSDART00000153482
|
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr4_-_9891874 | 0.08 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr3_+_13624815 | 0.07 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr1_+_47165842 | 0.07 |
ENSDART00000053152
ENSDART00000167051 |
cbr1
|
carbonyl reductase 1 |
| chr12_-_42214 | 0.07 |
ENSDART00000045071
|
foxk2
|
forkhead box K2 |
| chr10_-_33297864 | 0.07 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr18_+_619619 | 0.07 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr11_-_45152702 | 0.07 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr11_-_45138857 | 0.07 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr15_+_44366556 | 0.07 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr10_+_16584382 | 0.07 |
ENSDART00000112039
|
CR790388.1
|
|
| chr9_-_32753535 | 0.07 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr13_+_27232848 | 0.07 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr20_+_25879826 | 0.07 |
ENSDART00000018519
|
zgc:153896
|
zgc:153896 |
| chr1_-_50859053 | 0.06 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr6_+_39232245 | 0.06 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr9_-_20372977 | 0.06 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr24_-_40744672 | 0.06 |
ENSDART00000160672
|
CU633479.1
|
|
| chr9_+_25776971 | 0.06 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr5_+_33287611 | 0.06 |
ENSDART00000125093
ENSDART00000146759 |
med22
|
mediator complex subunit 22 |
| chr21_+_35041399 | 0.06 |
ENSDART00000188183
ENSDART00000021645 |
kdelc2
|
KDEL (Lys-Asp-Glu-Leu) containing 2 |
| chr13_+_29462249 | 0.06 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr20_-_46362606 | 0.06 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr15_-_22074315 | 0.06 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr2_-_11258547 | 0.05 |
ENSDART00000165803
ENSDART00000193817 |
slc44a5a
|
solute carrier family 44, member 5a |
| chr19_+_43297546 | 0.05 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr21_-_7035599 | 0.05 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr11_+_45323455 | 0.05 |
ENSDART00000189249
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr15_-_21877726 | 0.05 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr1_+_31110817 | 0.05 |
ENSDART00000137863
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr25_+_5068442 | 0.05 |
ENSDART00000097522
|
parvg
|
parvin, gamma |
| chr9_-_6372535 | 0.05 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr24_-_31843173 | 0.05 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr17_-_45386546 | 0.05 |
ENSDART00000182647
|
tmem206
|
transmembrane protein 206 |
| chr9_+_21146862 | 0.05 |
ENSDART00000136365
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr18_+_24921587 | 0.05 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr10_+_2799285 | 0.05 |
ENSDART00000030709
|
pnx
|
posterior neuron-specific homeobox |
| chr8_+_54274355 | 0.05 |
ENSDART00000067639
|
TMCC1 (1 of many)
|
transmembrane and coiled-coil domain family 1 |
| chr11_+_36665359 | 0.05 |
ENSDART00000166144
|
si:ch211-11c3.9
|
si:ch211-11c3.9 |
| chr20_+_42978499 | 0.05 |
ENSDART00000138793
|
si:ch211-203k16.3
|
si:ch211-203k16.3 |
| chr2_+_33326522 | 0.04 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr19_-_42571829 | 0.04 |
ENSDART00000102606
|
zgc:103438
|
zgc:103438 |
| chr6_-_46941245 | 0.04 |
ENSDART00000037875
|
igfn1.1
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 1 |
| chr14_-_21959712 | 0.04 |
ENSDART00000021417
|
p2rx3a
|
purinergic receptor P2X, ligand-gated ion channel, 3a |
| chr25_-_26758253 | 0.04 |
ENSDART00000123004
|
si:dkeyp-73b11.8
|
si:dkeyp-73b11.8 |
| chr20_-_35040041 | 0.04 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr2_-_10600950 | 0.04 |
ENSDART00000160216
|
BX323564.1
|
|
| chr8_-_52229462 | 0.03 |
ENSDART00000185949
ENSDART00000051825 |
tcf7l1b
|
transcription factor 7 like 1b |
| chr8_+_23521974 | 0.03 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr21_-_37973819 | 0.03 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr16_+_31804590 | 0.03 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr20_-_47188966 | 0.03 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr19_+_30884706 | 0.03 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr22_-_8725768 | 0.03 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr10_+_31646020 | 0.03 |
ENSDART00000115251
|
esama
|
endothelial cell adhesion molecule a |
| chr5_+_37840914 | 0.03 |
ENSDART00000097738
|
panx1b
|
pannexin 1b |
| chr20_-_40755614 | 0.02 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr4_-_15103646 | 0.02 |
ENSDART00000138183
ENSDART00000181044 |
nrf1
|
nuclear respiratory factor 1 |
| chr5_+_4436405 | 0.02 |
ENSDART00000167969
|
CABZ01079241.1
|
|
| chr17_-_23416897 | 0.02 |
ENSDART00000163391
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr3_+_1637358 | 0.02 |
ENSDART00000180266
|
CR394546.5
|
|
| chr13_-_20381485 | 0.02 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr13_+_35637048 | 0.02 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr17_+_53425037 | 0.02 |
ENSDART00000154983
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr7_+_2236317 | 0.02 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr22_+_19366866 | 0.02 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr20_+_12830448 | 0.02 |
ENSDART00000164754
|
BX547930.4
|
|
| chr2_-_22927958 | 0.02 |
ENSDART00000141621
|
myo7bb
|
myosin VIIBb |
| chr1_-_57280585 | 0.02 |
ENSDART00000152220
|
si:dkey-27j5.5
|
si:dkey-27j5.5 |
| chr22_-_392224 | 0.02 |
ENSDART00000124720
|
CABZ01072989.1
|
|
| chr1_-_47071979 | 0.02 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr4_+_11479705 | 0.02 |
ENSDART00000019458
ENSDART00000150587 ENSDART00000139370 ENSDART00000135826 |
asb13a.1
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 1 |
| chr22_+_20427170 | 0.02 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr12_+_16168342 | 0.02 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr20_-_9095105 | 0.02 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr24_+_9412450 | 0.02 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr7_+_48667081 | 0.02 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr4_+_69559692 | 0.02 |
ENSDART00000164383
|
znf993
|
zinc finger protein 993 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gsx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 0.2 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.1 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.2 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.2 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.5 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.1 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.7 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.0 | 0.1 | GO:0032263 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.1 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.1 | GO:0098543 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.0 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.0 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.2 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.4 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 0.2 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.1 | GO:0097300 | necrotic cell death(GO:0070265) cellular response to hydrogen peroxide(GO:0070301) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.2 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.2 | GO:0032143 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.3 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |