Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
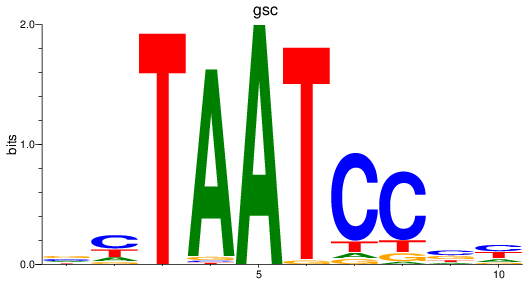
Results for gsc
Z-value: 0.61
Transcription factors associated with gsc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gsc
|
ENSDARG00000059073 | goosecoid |
|
gsc
|
ENSDARG00000111184 | goosecoid |
|
gsc
|
ENSDARG00000115937 | goosecoid |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gsc | dr11_v1_chr17_-_19345521_19345521 | -0.69 | 1.6e-03 | Click! |
Activity profile of gsc motif
Sorted Z-values of gsc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_2841205 | 2.09 |
ENSDART00000131505
ENSDART00000055869 |
ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr22_-_22147375 | 1.39 |
ENSDART00000149304
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr1_-_44928987 | 1.34 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr17_+_16090436 | 1.33 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr21_-_9562272 | 1.29 |
ENSDART00000162225
ENSDART00000163874 ENSDART00000168173 |
ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr18_+_49969568 | 1.22 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr9_+_8396755 | 1.21 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr8_+_2656231 | 1.19 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr8_+_2656681 | 1.18 |
ENSDART00000185067
ENSDART00000165943 |
fam102aa
|
family with sequence similarity 102, member Aa |
| chr2_-_58075414 | 1.18 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr19_+_24068223 | 1.16 |
ENSDART00000141351
ENSDART00000100420 |
pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr22_-_20695237 | 1.10 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr21_+_15756549 | 1.07 |
ENSDART00000026903
|
ptpra
|
protein tyrosine phosphatase, receptor type, A |
| chr17_+_35097024 | 1.01 |
ENSDART00000026152
|
asap2a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2a |
| chr7_+_66884291 | 1.00 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr14_-_45551572 | 0.99 |
ENSDART00000111410
|
ganab
|
glucosidase, alpha; neutral AB |
| chr21_-_20945658 | 0.99 |
ENSDART00000079701
|
rnf180
|
ring finger protein 180 |
| chr13_-_36525982 | 0.97 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr2_-_57264262 | 0.96 |
ENSDART00000183815
ENSDART00000149829 ENSDART00000088508 ENSDART00000149508 |
mbd3a
|
methyl-CpG binding domain protein 3a |
| chr5_+_25733774 | 0.93 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr19_-_11846958 | 0.89 |
ENSDART00000148516
|
ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr2_-_32262287 | 0.86 |
ENSDART00000056621
ENSDART00000039717 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr18_+_6638726 | 0.84 |
ENSDART00000142755
ENSDART00000167781 |
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr23_-_17509656 | 0.83 |
ENSDART00000148423
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr17_-_12249990 | 0.82 |
ENSDART00000177889
ENSDART00000155545 |
ahctf1
|
AT hook containing transcription factor 1 |
| chr6_-_17999776 | 0.81 |
ENSDART00000183048
ENSDART00000181577 ENSDART00000170597 |
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr14_-_25928899 | 0.81 |
ENSDART00000143518
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr8_-_25814263 | 0.81 |
ENSDART00000143397
|
taf10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr7_+_66884570 | 0.81 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr18_+_6638974 | 0.80 |
ENSDART00000162398
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr24_-_24060460 | 0.76 |
ENSDART00000142813
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr20_-_25643667 | 0.76 |
ENSDART00000137457
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr22_+_438714 | 0.71 |
ENSDART00000136491
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr19_-_6193067 | 0.71 |
ENSDART00000092656
ENSDART00000140347 |
erf
|
Ets2 repressor factor |
| chr5_+_60518611 | 0.70 |
ENSDART00000130565
ENSDART00000186310 |
tmem132e
|
transmembrane protein 132E |
| chrM_+_12897 | 0.70 |
ENSDART00000093622
|
mt-nd5
|
NADH dehydrogenase 5, mitochondrial |
| chr1_+_16574312 | 0.70 |
ENSDART00000187067
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr3_+_46764278 | 0.69 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr24_-_24060632 | 0.69 |
ENSDART00000090514
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr14_+_7699443 | 0.68 |
ENSDART00000123139
|
brd8
|
bromodomain containing 8 |
| chr14_-_25935167 | 0.68 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr22_+_34701848 | 0.67 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr16_-_20932896 | 0.65 |
ENSDART00000180646
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr5_+_59030096 | 0.65 |
ENSDART00000179814
ENSDART00000110182 |
rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr3_-_49504023 | 0.64 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr20_-_34663209 | 0.64 |
ENSDART00000132545
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr7_-_9674073 | 0.64 |
ENSDART00000187902
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr3_-_23513155 | 0.63 |
ENSDART00000170200
|
BX682558.1
|
|
| chr2_-_32513538 | 0.63 |
ENSDART00000056640
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr5_-_24029228 | 0.62 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr15_+_45591669 | 0.61 |
ENSDART00000157459
|
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr17_-_8656155 | 0.59 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr8_+_21280360 | 0.58 |
ENSDART00000144488
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr11_+_31680513 | 0.58 |
ENSDART00000139900
ENSDART00000040305 |
diaph3
|
diaphanous-related formin 3 |
| chr17_-_33414781 | 0.58 |
ENSDART00000142203
ENSDART00000034638 |
ccdc28a
|
coiled-coil domain containing 28A |
| chr16_-_22585289 | 0.57 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr15_-_26887028 | 0.56 |
ENSDART00000156292
|
si:dkey-243i1.1
|
si:dkey-243i1.1 |
| chr20_-_24182689 | 0.55 |
ENSDART00000171184
|
map3k7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr20_+_34671386 | 0.54 |
ENSDART00000152836
ENSDART00000138226 |
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr12_+_5048044 | 0.53 |
ENSDART00000161548
ENSDART00000172607 |
kif22
|
kinesin family member 22 |
| chr3_+_46764022 | 0.51 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr2_-_32387441 | 0.51 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr1_+_27690 | 0.51 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr23_-_31060350 | 0.50 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr6_+_12527725 | 0.50 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr18_+_21951551 | 0.50 |
ENSDART00000146261
|
ranbp10
|
RAN binding protein 10 |
| chr2_-_30784198 | 0.50 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr24_-_24271629 | 0.50 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr20_+_32473584 | 0.49 |
ENSDART00000192449
|
ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr24_+_3478871 | 0.47 |
ENSDART00000111491
ENSDART00000134598 ENSDART00000142407 |
wdr37
|
WD repeat domain 37 |
| chr24_+_37533728 | 0.46 |
ENSDART00000061203
|
rhot2
|
ras homolog family member T2 |
| chr5_+_31791001 | 0.46 |
ENSDART00000043010
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr9_-_52598343 | 0.45 |
ENSDART00000167922
|
xrcc5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 |
| chr7_-_49594995 | 0.45 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr12_+_33460794 | 0.44 |
ENSDART00000007053
ENSDART00000142716 |
narf
|
nuclear prelamin A recognition factor |
| chr2_-_9696283 | 0.41 |
ENSDART00000165712
|
selenot1a
|
selenoprotein T, 1a |
| chr3_-_29891456 | 0.40 |
ENSDART00000151677
ENSDART00000014021 |
slc25a39
|
solute carrier family 25, member 39 |
| chr19_+_3140313 | 0.39 |
ENSDART00000125504
|
zgc:86598
|
zgc:86598 |
| chr16_+_11297703 | 0.39 |
ENSDART00000125158
|
znf574
|
zinc finger protein 574 |
| chr5_+_47882319 | 0.39 |
ENSDART00000149316
|
rasa1a
|
RAS p21 protein activator (GTPase activating protein) 1a |
| chr12_-_19119176 | 0.37 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr2_+_30531726 | 0.36 |
ENSDART00000146518
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr25_-_32381642 | 0.36 |
ENSDART00000133872
ENSDART00000006124 |
cops2
|
COP9 signalosome subunit 2 |
| chr6_-_19270484 | 0.36 |
ENSDART00000186894
ENSDART00000188709 |
zgc:174863
|
zgc:174863 |
| chr19_+_18797623 | 0.36 |
ENSDART00000166172
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr14_+_30753666 | 0.36 |
ENSDART00000009385
ENSDART00000137782 ENSDART00000172778 |
yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr7_+_17063761 | 0.35 |
ENSDART00000182880
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr25_-_22889519 | 0.35 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr1_+_16573982 | 0.35 |
ENSDART00000166317
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr3_+_14157090 | 0.35 |
ENSDART00000156766
|
si:ch211-108d22.2
|
si:ch211-108d22.2 |
| chr20_+_37866686 | 0.34 |
ENSDART00000036546
|
vash2
|
vasohibin 2 |
| chr24_-_7777389 | 0.33 |
ENSDART00000138541
|
rpgrip1
|
RPGR interacting protein 1 |
| chr23_+_17509794 | 0.32 |
ENSDART00000148457
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr6_-_43283122 | 0.32 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr19_-_32944050 | 0.32 |
ENSDART00000137611
|
azin1b
|
antizyme inhibitor 1b |
| chr3_-_29891218 | 0.31 |
ENSDART00000142118
|
slc25a39
|
solute carrier family 25, member 39 |
| chr21_-_28235361 | 0.31 |
ENSDART00000164458
|
nrxn2a
|
neurexin 2a |
| chr14_+_32838110 | 0.31 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr7_-_57933736 | 0.30 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr23_-_27152866 | 0.30 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr20_+_26987416 | 0.30 |
ENSDART00000012816
|
sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr10_+_6013076 | 0.30 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr1_+_41690402 | 0.29 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr18_-_38245062 | 0.29 |
ENSDART00000189092
|
nat10
|
N-acetyltransferase 10 |
| chr13_+_39182099 | 0.28 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr20_-_20270191 | 0.28 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr20_+_37866861 | 0.27 |
ENSDART00000153220
|
vash2
|
vasohibin 2 |
| chr8_-_48847772 | 0.27 |
ENSDART00000122458
|
wrap73
|
WD repeat containing, antisense to TP73 |
| chr17_-_2685026 | 0.27 |
ENSDART00000191014
ENSDART00000179309 |
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr6_-_59381391 | 0.27 |
ENSDART00000157066
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr23_-_25779995 | 0.26 |
ENSDART00000110670
|
si:dkey-21c19.3
|
si:dkey-21c19.3 |
| chr24_-_16905018 | 0.26 |
ENSDART00000066759
|
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr11_-_15090118 | 0.25 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr10_-_41400049 | 0.25 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr5_-_57526807 | 0.25 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr23_+_39854566 | 0.25 |
ENSDART00000190423
ENSDART00000164473 ENSDART00000161881 |
si:ch73-217b7.1
|
si:ch73-217b7.1 |
| chr23_+_6232895 | 0.25 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr18_-_21640389 | 0.23 |
ENSDART00000100857
|
slc38a8a
|
solute carrier family 38, member 8a |
| chr24_+_9881219 | 0.23 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr19_+_18799319 | 0.22 |
ENSDART00000171843
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr14_+_32837914 | 0.22 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr6_-_33924883 | 0.22 |
ENSDART00000132762
ENSDART00000148142 ENSDART00000142213 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr24_+_37825634 | 0.21 |
ENSDART00000129889
|
ift140
|
intraflagellar transport 140 homolog (Chlamydomonas) |
| chr7_-_21905851 | 0.20 |
ENSDART00000111066
ENSDART00000020288 |
epoa
|
erythropoietin a |
| chr15_+_42995381 | 0.20 |
ENSDART00000152222
|
acsl3a
|
acyl-CoA synthetase long chain family member 3a |
| chr14_+_32839535 | 0.20 |
ENSDART00000168975
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr25_+_7299488 | 0.19 |
ENSDART00000184836
|
hmg20a
|
high mobility group 20A |
| chr21_+_33187992 | 0.19 |
ENSDART00000162745
ENSDART00000188388 |
BX072577.1
|
|
| chr23_-_10137322 | 0.19 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr5_+_26199537 | 0.18 |
ENSDART00000144303
|
zgc:86811
|
zgc:86811 |
| chr17_-_14559576 | 0.18 |
ENSDART00000162452
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr3_-_10970502 | 0.18 |
ENSDART00000127500
|
CR382337.1
|
|
| chr17_-_50010121 | 0.16 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr24_-_16904803 | 0.15 |
ENSDART00000165972
ENSDART00000149974 ENSDART00000149706 |
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr14_+_21754521 | 0.14 |
ENSDART00000111839
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr18_-_25646286 | 0.14 |
ENSDART00000099511
ENSDART00000186890 |
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr19_+_2631565 | 0.14 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr10_+_36345176 | 0.14 |
ENSDART00000099397
|
or105-1
|
odorant receptor, family C, subfamily 105, member 1 |
| chr7_+_52887701 | 0.14 |
ENSDART00000109973
|
tp53bp1
|
tumor protein p53 binding protein, 1 |
| chr1_-_45177373 | 0.14 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr11_-_15090564 | 0.13 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr2_+_4146299 | 0.13 |
ENSDART00000173418
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr21_-_2348838 | 0.12 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr20_+_29587995 | 0.12 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr23_-_1587955 | 0.11 |
ENSDART00000136037
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr20_-_18736281 | 0.11 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr22_+_661505 | 0.10 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr10_-_25628555 | 0.10 |
ENSDART00000143978
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr14_+_44545092 | 0.10 |
ENSDART00000175454
|
lingo2a
|
leucine rich repeat and Ig domain containing 2a |
| chr4_+_33012407 | 0.08 |
ENSDART00000151873
|
si:dkey-26h11.2
|
si:dkey-26h11.2 |
| chr2_+_4146606 | 0.07 |
ENSDART00000171170
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr23_+_13814978 | 0.07 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr3_-_5277041 | 0.07 |
ENSDART00000046995
|
txn2
|
thioredoxin 2 |
| chr3_-_36209936 | 0.06 |
ENSDART00000175208
|
csnk1da
|
casein kinase 1, delta a |
| chr25_+_16079913 | 0.06 |
ENSDART00000146350
|
far1
|
fatty acyl CoA reductase 1 |
| chr8_-_54077740 | 0.05 |
ENSDART00000027000
|
rho
|
rhodopsin |
| chr2_-_16986894 | 0.05 |
ENSDART00000145720
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr15_+_9053059 | 0.05 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr5_-_14390445 | 0.05 |
ENSDART00000026120
|
ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr11_-_29563437 | 0.04 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr2_+_36828746 | 0.04 |
ENSDART00000186638
|
nrd1a
|
nardilysin a (N-arginine dibasic convertase) |
| chr17_-_2690083 | 0.04 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr17_-_51202339 | 0.04 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr19_+_4061699 | 0.04 |
ENSDART00000158309
ENSDART00000166512 |
btr25
btr26
|
bloodthirsty-related gene family, member 25 bloodthirsty-related gene family, member 26 |
| chr7_+_30970045 | 0.04 |
ENSDART00000155974
|
tjp1a
|
tight junction protein 1a |
| chr7_-_66864756 | 0.04 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr22_+_38581012 | 0.03 |
ENSDART00000169239
|
BX088728.1
|
|
| chr1_-_11627289 | 0.03 |
ENSDART00000163312
|
si:dkey-26i13.3
|
si:dkey-26i13.3 |
| chr17_-_15657029 | 0.03 |
ENSDART00000153925
|
fut9a
|
fucosyltransferase 9a |
| chr23_-_18981595 | 0.03 |
ENSDART00000147617
|
bcl2l1
|
bcl2-like 1 |
| chr9_+_20519846 | 0.03 |
ENSDART00000109680
ENSDART00000142220 |
vtcn1
|
V-set domain containing T cell activation inhibitor 1 |
| chr18_+_7639401 | 0.03 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr15_+_45640906 | 0.03 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr1_-_45146834 | 0.03 |
ENSDART00000144997
|
si:ch211-239f4.6
|
si:ch211-239f4.6 |
| chr22_+_5103349 | 0.02 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr3_-_33574576 | 0.02 |
ENSDART00000184881
|
CR847537.1
|
|
| chr9_-_29544720 | 0.02 |
ENSDART00000130317
|
arhgap20
|
Rho GTPase activating protein 20 |
| chr19_+_42693855 | 0.02 |
ENSDART00000136873
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr16_+_29555801 | 0.02 |
ENSDART00000169425
|
ensab
|
endosulfine alpha b |
| chr11_+_36180349 | 0.02 |
ENSDART00000012940
|
grm2b
|
glutamate receptor, metabotropic 2b |
| chr25_+_16080181 | 0.01 |
ENSDART00000061753
|
far1
|
fatty acyl CoA reductase 1 |
| chr7_-_17780048 | 0.01 |
ENSDART00000183336
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr24_+_5826005 | 0.01 |
ENSDART00000154930
|
si:ch211-157j23.5
|
si:ch211-157j23.5 |
| chr14_+_33486179 | 0.01 |
ENSDART00000149254
|
cul4b
|
cullin 4B |
| chr2_+_29795833 | 0.01 |
ENSDART00000056750
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr9_+_21990095 | 0.01 |
ENSDART00000146829
ENSDART00000133515 ENSDART00000193582 |
si:dkey-57a22.13
|
si:dkey-57a22.13 |
| chr7_-_18656069 | 0.01 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr2_+_36828243 | 0.01 |
ENSDART00000135754
ENSDART00000140474 |
nrd1a
|
nardilysin a (N-arginine dibasic convertase) |
| chr17_-_32426392 | 0.01 |
ENSDART00000148455
ENSDART00000149885 ENSDART00000179314 |
grhl1
|
grainyhead-like transcription factor 1 |
| chr1_-_11596829 | 0.00 |
ENSDART00000140725
|
si:dkey-26i13.7
|
si:dkey-26i13.7 |
| chr16_+_19536614 | 0.00 |
ENSDART00000112894
ENSDART00000079201 ENSDART00000139357 |
sp8b
|
sp8 transcription factor b |
| chr10_+_13279079 | 0.00 |
ENSDART00000135082
|
tmem267
|
transmembrane protein 267 |
| chr1_-_11616142 | 0.00 |
ENSDART00000131668
|
si:dkey-26i13.5
|
si:dkey-26i13.5 |
| chr15_-_38154616 | 0.00 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gsc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.3 | 1.6 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.3 | 0.9 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 1.0 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.2 | 1.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.2 | 0.5 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.2 | 0.7 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.2 | 0.8 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.6 | GO:0045428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.1 | 0.6 | GO:0039703 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.3 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.6 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.9 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.6 | GO:0071331 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 | 0.2 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.1 | 0.7 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 0.7 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.7 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 1.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.3 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.5 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 1.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.8 | GO:0071230 | TORC1 signaling(GO:0038202) cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.6 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.5 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.6 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 0.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.3 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.5 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.1 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.3 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 1.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.0 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.4 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.1 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.4 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.1 | 1.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 1.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.7 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 0.7 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.3 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.1 | 0.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.9 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.1 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.9 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |