Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for grhl1
Z-value: 0.65
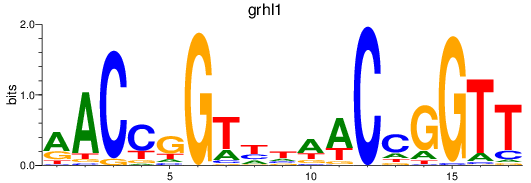
Transcription factors associated with grhl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
grhl1
|
ENSDARG00000061391 | grainyhead-like transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| grhl1 | dr11_v1_chr17_-_32426392_32426413 | 0.85 | 7.7e-06 | Click! |
Activity profile of grhl1 motif
Sorted Z-values of grhl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_9780931 | 1.93 |
ENSDART00000134280
ENSDART00000150664 ENSDART00000150304 ENSDART00000080744 |
svopl
|
SVOP-like |
| chr22_-_22337382 | 1.78 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr2_-_58075414 | 1.59 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr2_-_47620806 | 1.52 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr1_-_9858508 | 1.45 |
ENSDART00000147904
|
mad1l1
|
mitotic arrest deficient 1 like 1 |
| chr25_-_24202576 | 1.38 |
ENSDART00000048507
|
uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr16_+_35905031 | 1.38 |
ENSDART00000162411
|
sh3d21
|
SH3 domain containing 21 |
| chr14_-_33945692 | 1.25 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr20_-_43346049 | 1.24 |
ENSDART00000152888
|
afdna
|
afadin, adherens junction formation factor a |
| chr4_-_14954327 | 1.21 |
ENSDART00000182729
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr10_-_36793412 | 1.18 |
ENSDART00000185966
|
dhrs13a.2
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 2 |
| chr9_-_30264415 | 1.18 |
ENSDART00000060150
|
mid1ip1a
|
MID1 interacting protein 1a |
| chr17_+_28102487 | 1.18 |
ENSDART00000131638
ENSDART00000076265 |
zgc:91908
|
zgc:91908 |
| chr1_-_31105376 | 1.16 |
ENSDART00000132466
|
ppp1r9alb
|
protein phosphatase 1 regulatory subunit 9A-like B |
| chr4_-_14954029 | 1.05 |
ENSDART00000038642
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr21_-_3700334 | 1.05 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr12_-_28794957 | 1.03 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr1_-_48933 | 1.01 |
ENSDART00000171162
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr14_-_763744 | 0.93 |
ENSDART00000165856
|
trim35-27
|
tripartite motif containing 35-27 |
| chr20_-_42378865 | 0.89 |
ENSDART00000139912
ENSDART00000015801 |
dcbld1
|
discoidin, CUB and LCCL domain containing 1 |
| chr3_+_32832042 | 0.83 |
ENSDART00000132679
ENSDART00000035759 |
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr5_+_43470544 | 0.83 |
ENSDART00000111587
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr15_-_5901514 | 0.81 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr23_+_43668756 | 0.80 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr3_-_30186296 | 0.77 |
ENSDART00000134395
ENSDART00000077057 ENSDART00000017422 |
tbc1d17
|
TBC1 domain family, member 17 |
| chr20_+_20731052 | 0.66 |
ENSDART00000047662
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr5_-_6567464 | 0.66 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr5_+_58665648 | 0.65 |
ENSDART00000167481
|
zgc:194948
|
zgc:194948 |
| chr3_-_32831429 | 0.61 |
ENSDART00000184932
|
zgc:153733
|
zgc:153733 |
| chr14_+_4276394 | 0.59 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr3_-_32831971 | 0.57 |
ENSDART00000075270
|
zgc:153733
|
zgc:153733 |
| chr16_+_23431189 | 0.53 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr16_+_53526135 | 0.51 |
ENSDART00000083558
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr7_+_54259657 | 0.45 |
ENSDART00000170174
|
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr15_+_42560354 | 0.44 |
ENSDART00000059484
|
CLDN8 (1 of many)
|
zgc:110333 |
| chr14_-_4044545 | 0.43 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr19_-_32500373 | 0.42 |
ENSDART00000052104
|
fuca1.1
|
alpha-L-fucosidase 1, tandem duplicate 1 |
| chr25_+_15994100 | 0.36 |
ENSDART00000144723
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr6_+_23057311 | 0.36 |
ENSDART00000026448
|
evpla
|
envoplakin a |
| chr1_+_49352900 | 0.33 |
ENSDART00000008468
|
msx1b
|
muscle segment homeobox 1b |
| chr5_+_4006837 | 0.30 |
ENSDART00000138862
|
pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr16_-_10837245 | 0.25 |
ENSDART00000036891
|
rabac1
|
Rab acceptor 1 (prenylated) |
| chr3_-_55511569 | 0.25 |
ENSDART00000108995
|
tex2
|
testis expressed 2 |
| chr16_+_35661771 | 0.25 |
ENSDART00000161393
|
map7d1a
|
MAP7 domain containing 1a |
| chr15_+_41919484 | 0.21 |
ENSDART00000099821
ENSDART00000146246 |
nlrp16
|
NACHT, LRR and PYD domains-containing protein 16 |
| chr4_+_76722754 | 0.19 |
ENSDART00000153867
|
ms4a17a.3
|
membrane-spanning 4-domains, subfamily A, member 17A.3 |
| chr16_+_35661331 | 0.19 |
ENSDART00000161725
|
map7d1a
|
MAP7 domain containing 1a |
| chr19_+_30885258 | 0.18 |
ENSDART00000143394
|
yars
|
tyrosyl-tRNA synthetase |
| chr25_+_7435291 | 0.13 |
ENSDART00000172567
ENSDART00000163017 |
prc1a
|
protein regulator of cytokinesis 1a |
| chr2_+_5521671 | 0.13 |
ENSDART00000099647
ENSDART00000138443 |
crfb16
|
cytokine receptor family member B16 |
| chr16_-_9675982 | 0.12 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr22_+_25672155 | 0.12 |
ENSDART00000087769
|
si:ch211-250e5.2
|
si:ch211-250e5.2 |
| chr23_-_22650668 | 0.12 |
ENSDART00000132733
|
ca6
|
carbonic anhydrase VI |
| chr15_-_12360409 | 0.12 |
ENSDART00000164596
|
tmprss13a
|
transmembrane protease, serine 13a |
| chr23_-_22650992 | 0.11 |
ENSDART00000079007
|
ca6
|
carbonic anhydrase VI |
| chr2_+_7557912 | 0.10 |
ENSDART00000160053
|
ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr8_+_3405612 | 0.08 |
ENSDART00000163437
|
zgc:112433
|
zgc:112433 |
| chr8_-_36399884 | 0.06 |
ENSDART00000108538
|
si:zfos-2070c2.3
|
si:zfos-2070c2.3 |
| chr5_-_57686487 | 0.05 |
ENSDART00000074264
|
crfb12
|
cytokine receptor family member B12 |
| chr2_-_42864472 | 0.05 |
ENSDART00000134139
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr11_-_45140227 | 0.05 |
ENSDART00000184062
|
cant1b
|
calcium activated nucleotidase 1b |
| chr3_+_45756605 | 0.04 |
ENSDART00000189772
|
gper1
|
G protein-coupled estrogen receptor 1 |
| chr15_+_41901710 | 0.04 |
ENSDART00000159874
|
si:ch211-191a16.5
|
si:ch211-191a16.5 |
| chr21_+_11244068 | 0.04 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr22_-_14361022 | 0.03 |
ENSDART00000140224
|
si:dkeyp-122a9.2
|
si:dkeyp-122a9.2 |
| chr15_+_41901970 | 0.03 |
ENSDART00000152724
|
si:ch211-191a16.5
|
si:ch211-191a16.5 |
| chr11_+_8152872 | 0.03 |
ENSDART00000091638
ENSDART00000138057 ENSDART00000166379 |
samd13
|
sterile alpha motif domain containing 13 |
| chr4_+_4232562 | 0.03 |
ENSDART00000177529
|
smkr1
|
small lysine rich protein 1 |
| chr4_-_26413391 | 0.03 |
ENSDART00000145955
|
b4galnt3a
|
beta-1,4-N-acetyl-galactosaminyl transferase 3a |
| chr5_-_37900350 | 0.02 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr6_+_30668098 | 0.01 |
ENSDART00000112294
|
ttc22
|
tetratricopeptide repeat domain 22 |
| chr12_+_30360579 | 0.01 |
ENSDART00000152900
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of grhl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.4 | 1.8 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 0.8 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.6 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.2 | 1.4 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 1.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 1.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.5 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 1.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.8 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 1.2 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.0 | GO:1903430 | negative regulation of cell maturation(GO:1903430) |
| 0.0 | 0.1 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 1.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.2 | GO:0032880 | regulation of protein localization(GO:0032880) |
| 0.0 | 0.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.5 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.5 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.4 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 2.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 1.4 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |