Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for glis2a+glis2b
Z-value: 0.31
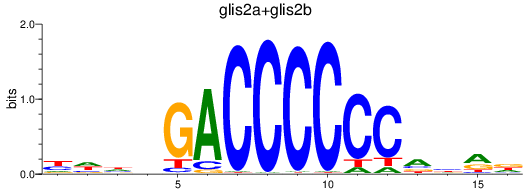
Transcription factors associated with glis2a+glis2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
glis2a
|
ENSDARG00000078388 | GLIS family zinc finger 2a |
|
glis2b
|
ENSDARG00000100232 | GLIS family zinc finger 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| glis2b | dr11_v1_chr3_+_12334509_12334509 | 0.36 | 1.5e-01 | Click! |
| glis2a | dr11_v1_chr22_-_26558166_26558166 | -0.21 | 4.0e-01 | Click! |
Activity profile of glis2a+glis2b motif
Sorted Z-values of glis2a+glis2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_10338554 | 1.11 |
ENSDART00000186936
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr17_-_4245311 | 1.02 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr6_-_7769178 | 0.85 |
ENSDART00000191701
ENSDART00000149823 |
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr12_+_22672323 | 0.81 |
ENSDART00000123808
ENSDART00000159864 |
cdca9
|
cell division cycle associated 9 |
| chr17_-_4245902 | 0.79 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr25_+_30196039 | 0.72 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr3_-_33113879 | 0.68 |
ENSDART00000044677
|
rarab
|
retinoic acid receptor, alpha b |
| chr4_-_1776352 | 0.62 |
ENSDART00000123089
|
dnajc2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr2_-_49031303 | 0.61 |
ENSDART00000143471
|
cdc34b
|
cell division cycle 34 homolog (S. cerevisiae) b |
| chr2_+_51783120 | 0.58 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr25_-_997894 | 0.57 |
ENSDART00000169011
|
znf609a
|
zinc finger protein 609a |
| chr3_+_40409100 | 0.47 |
ENSDART00000103486
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr7_-_24838857 | 0.42 |
ENSDART00000179766
ENSDART00000180892 |
fam113
|
family with sequence similarity 113 |
| chr25_+_16912243 | 0.41 |
ENSDART00000186579
|
zgc:77158
|
zgc:77158 |
| chr5_+_25760112 | 0.41 |
ENSDART00000088011
ENSDART00000182046 |
tmem2
|
transmembrane protein 2 |
| chr19_+_15443540 | 0.36 |
ENSDART00000193355
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr23_+_43668756 | 0.35 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr23_-_31060350 | 0.34 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr12_-_41619257 | 0.34 |
ENSDART00000162967
|
dpysl4
|
dihydropyrimidinase-like 4 |
| chr15_+_518092 | 0.33 |
ENSDART00000154594
|
pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr13_+_1928361 | 0.31 |
ENSDART00000164764
|
hmgcll1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr1_+_15258641 | 0.31 |
ENSDART00000033018
|
pi4k2b
|
phosphatidylinositol 4-kinase type 2 beta |
| chr10_-_1961576 | 0.30 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr8_-_20914829 | 0.29 |
ENSDART00000025356
|
haus5
|
HAUS augmin-like complex, subunit 5 |
| chr6_+_38845848 | 0.26 |
ENSDART00000184907
|
stk35l
|
serine/threonine kinase 35, like |
| chr23_-_36934944 | 0.26 |
ENSDART00000109976
ENSDART00000162179 |
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr6_+_38845697 | 0.23 |
ENSDART00000053187
|
stk35l
|
serine/threonine kinase 35, like |
| chr2_-_9744081 | 0.22 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr2_+_24867534 | 0.20 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr13_+_39208542 | 0.20 |
ENSDART00000147971
|
fam135a
|
family with sequence similarity 135, member A |
| chr24_-_32582378 | 0.16 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr5_+_38886499 | 0.16 |
ENSDART00000076845
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr3_+_21189766 | 0.14 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr4_-_4756349 | 0.11 |
ENSDART00000057507
|
ssbp1
|
single-stranded DNA binding protein 1 |
| chr13_-_24880525 | 0.11 |
ENSDART00000136624
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr6_+_37625787 | 0.10 |
ENSDART00000065122
|
tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr24_-_35707552 | 0.09 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr19_-_42928979 | 0.09 |
ENSDART00000086659
|
cyhr1
|
cysteine/histidine-rich 1 |
| chr4_-_59152337 | 0.09 |
ENSDART00000156770
|
znf1149
|
zinc finger protein 1149 |
| chr7_+_11390462 | 0.08 |
ENSDART00000114383
|
tlnrd1
|
talin rod domain containing 1 |
| chr23_+_39481537 | 0.07 |
ENSDART00000187222
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr19_-_36640677 | 0.05 |
ENSDART00000189743
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr9_+_20781047 | 0.04 |
ENSDART00000139174
|
fam46c
|
family with sequence similarity 46, member C |
| chr19_-_48336535 | 0.04 |
ENSDART00000162752
|
si:ch73-359m17.6
|
si:ch73-359m17.6 |
| chr23_-_26536055 | 0.03 |
ENSDART00000182719
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr22_-_18116635 | 0.03 |
ENSDART00000005724
|
ncanb
|
neurocan b |
| chr3_-_58078522 | 0.02 |
ENSDART00000154775
|
si:ch211-256e16.8
|
si:ch211-256e16.8 |
| chr7_-_19642417 | 0.02 |
ENSDART00000160936
|
si:ch211-212k18.4
|
si:ch211-212k18.4 |
| chr24_+_25471196 | 0.02 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr20_+_38285671 | 0.02 |
ENSDART00000061432
|
ccl38a.4
|
chemokine (C-C motif) ligand 38, duplicate 4 |
| chr5_-_71995108 | 0.02 |
ENSDART00000124587
|
fam78ab
|
family with sequence similarity 78, member Ab |
| chr18_+_17151916 | 0.01 |
ENSDART00000175488
|
trhr2
|
thyrotropin releasing hormone receptor 2 |
| chr9_+_54158387 | 0.01 |
ENSDART00000171315
ENSDART00000180570 |
TLR8
|
toll like receptor 8 |
| chr10_+_21701568 | 0.01 |
ENSDART00000090748
|
pcdh1g9
|
protocadherin 1 gamma 9 |
| chr2_+_13722116 | 0.00 |
ENSDART00000155015
|
zbtb41
|
zinc finger and BTB domain containing 41 |
| chr16_-_26435431 | 0.00 |
ENSDART00000187526
|
megf8
|
multiple EGF-like-domains 8 |
| chr17_-_44968177 | 0.00 |
ENSDART00000075510
|
ngb
|
neuroglobin |
Network of associatons between targets according to the STRING database.
First level regulatory network of glis2a+glis2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.2 | 0.8 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.3 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.1 | 0.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.4 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.7 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.1 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.2 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.3 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.4 | GO:0015154 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.3 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.8 | GO:0008083 | growth factor activity(GO:0008083) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |