Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
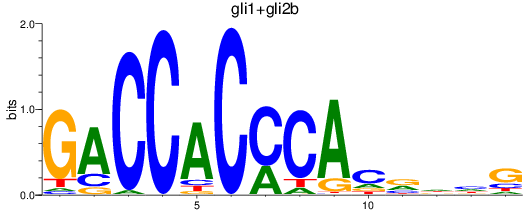
Results for gli1+gli2b
Z-value: 0.49
Transcription factors associated with gli1+gli2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gli2b
|
ENSDARG00000020884 | GLI family zinc finger 2b |
|
gli1
|
ENSDARG00000101244 | GLI family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gli1 | dr11_v1_chr6_-_59505589_59505589 | -0.85 | 7.9e-06 | Click! |
| gli2b | dr11_v1_chr11_+_43661735_43661735 | 0.54 | 2.1e-02 | Click! |
Activity profile of gli1+gli2b motif
Sorted Z-values of gli1+gli2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_15155684 | 1.06 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr7_-_4110462 | 1.03 |
ENSDART00000173318
|
zgc:55733
|
zgc:55733 |
| chr20_-_41992878 | 1.00 |
ENSDART00000100967
|
si:dkeyp-114g9.1
|
si:dkeyp-114g9.1 |
| chr24_-_17029374 | 0.96 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr9_+_45227028 | 0.83 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr22_-_38459316 | 0.76 |
ENSDART00000149683
ENSDART00000098461 |
ptk7a
|
protein tyrosine kinase 7a |
| chr13_-_33317323 | 0.76 |
ENSDART00000110295
ENSDART00000144848 ENSDART00000136701 |
tmem234
|
transmembrane protein 234 |
| chr10_+_16036573 | 0.75 |
ENSDART00000188757
|
lmnb1
|
lamin B1 |
| chr15_+_25489406 | 0.75 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr18_-_158541 | 0.74 |
ENSDART00000188914
ENSDART00000191052 |
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr22_-_22340688 | 0.73 |
ENSDART00000105597
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr22_-_22147375 | 0.72 |
ENSDART00000149304
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr10_+_16036246 | 0.72 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr1_+_54737353 | 0.72 |
ENSDART00000130675
ENSDART00000162075 |
pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr18_-_40753583 | 0.70 |
ENSDART00000026767
|
akt2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr11_+_5468629 | 0.68 |
ENSDART00000013203
|
cse1l
|
CSE1 chromosome segregation 1-like (yeast) |
| chr19_-_8926575 | 0.67 |
ENSDART00000080897
|
rprd2a
|
regulation of nuclear pre-mRNA domain containing 2a |
| chr14_+_28486213 | 0.64 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr17_-_4252221 | 0.62 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr1_-_14506759 | 0.61 |
ENSDART00000057044
|
si:dkey-194g4.1
|
si:dkey-194g4.1 |
| chr2_-_57707039 | 0.61 |
ENSDART00000097685
|
CABZ01060891.1
|
|
| chr19_+_7424347 | 0.60 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr24_+_39641991 | 0.56 |
ENSDART00000142182
|
luc7l
|
LUC7-like (S. cerevisiae) |
| chr5_+_13472234 | 0.56 |
ENSDART00000114069
ENSDART00000132406 |
cnnm4b
|
cyclin and CBS domain divalent metal cation transport mediator 4b |
| chr20_+_51730658 | 0.54 |
ENSDART00000010271
|
aida
|
axin interactor, dorsalization associated |
| chr14_-_15155384 | 0.54 |
ENSDART00000172666
|
uvssa
|
UV-stimulated scaffold protein A |
| chr15_+_24737599 | 0.52 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr2_+_51783120 | 0.51 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr1_+_19434198 | 0.51 |
ENSDART00000012552
|
clockb
|
clock circadian regulator b |
| chr7_+_10701938 | 0.49 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr19_-_46566430 | 0.49 |
ENSDART00000166668
|
ext1b
|
exostosin glycosyltransferase 1b |
| chr1_-_9527200 | 0.49 |
ENSDART00000110790
|
si:ch73-12o23.1
|
si:ch73-12o23.1 |
| chr19_+_48060464 | 0.48 |
ENSDART00000123163
|
zgc:85936
|
zgc:85936 |
| chr25_+_14697247 | 0.48 |
ENSDART00000180747
|
mpped2
|
metallophosphoesterase domain containing 2b |
| chr7_+_10701770 | 0.47 |
ENSDART00000167323
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr12_+_13091842 | 0.46 |
ENSDART00000185477
ENSDART00000181435 ENSDART00000124799 |
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr23_+_4226341 | 0.45 |
ENSDART00000012445
|
zgc:113278
|
zgc:113278 |
| chr6_+_27090800 | 0.45 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr24_+_39518774 | 0.45 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr21_-_17037907 | 0.44 |
ENSDART00000101263
|
ube2g1b
|
ubiquitin-conjugating enzyme E2G 1b (UBC7 homolog, yeast) |
| chr20_-_51186524 | 0.44 |
ENSDART00000027836
ENSDART00000114407 |
rbm25b
|
RNA binding motif protein 25b |
| chr5_+_66326004 | 0.43 |
ENSDART00000144351
|
malt1
|
MALT paracaspase 1 |
| chr13_+_7242916 | 0.43 |
ENSDART00000184238
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr22_+_26665422 | 0.42 |
ENSDART00000164994
|
adcy9
|
adenylate cyclase 9 |
| chr19_+_9050852 | 0.42 |
ENSDART00000151031
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr16_+_13822137 | 0.42 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr13_-_865193 | 0.41 |
ENSDART00000187053
|
AL929536.7
|
|
| chr9_+_4252839 | 0.40 |
ENSDART00000169740
|
kalrna
|
kalirin RhoGEF kinase a |
| chr3_+_34986837 | 0.39 |
ENSDART00000190341
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr25_-_34740627 | 0.39 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr13_+_15657911 | 0.38 |
ENSDART00000134972
ENSDART00000138991 ENSDART00000133342 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr17_-_17447899 | 0.38 |
ENSDART00000156928
ENSDART00000109034 |
nrxn3a
|
neurexin 3a |
| chr13_-_1423008 | 0.38 |
ENSDART00000110828
|
znf451
|
zinc finger protein 451 |
| chr14_+_49233896 | 0.36 |
ENSDART00000159526
|
rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| chr11_+_24314148 | 0.34 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr13_-_50002852 | 0.34 |
ENSDART00000099439
|
lyst
|
lysosomal trafficking regulator |
| chr5_+_52844681 | 0.34 |
ENSDART00000162459
ENSDART00000184914 |
scarb2a
|
scavenger receptor class B, member 2a |
| chr3_-_33941319 | 0.34 |
ENSDART00000026090
ENSDART00000111878 |
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr9_-_46072805 | 0.33 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr3_+_45365098 | 0.33 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr24_+_39137001 | 0.32 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr17_+_34215886 | 0.32 |
ENSDART00000186775
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr5_-_17876709 | 0.31 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr14_-_16476863 | 0.30 |
ENSDART00000089021
|
canx
|
calnexin |
| chr2_+_54086436 | 0.29 |
ENSDART00000174581
|
CU179656.1
|
|
| chr15_-_19128705 | 0.28 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr11_+_2855430 | 0.28 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr5_-_57311037 | 0.27 |
ENSDART00000149855
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr15_-_43287515 | 0.26 |
ENSDART00000155103
|
prss16
|
protease, serine, 16 (thymus) |
| chr3_+_45368973 | 0.26 |
ENSDART00000187282
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr6_-_3982783 | 0.24 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr2_-_30784198 | 0.24 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr3_-_21062706 | 0.24 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr9_-_10145795 | 0.23 |
ENSDART00000004745
ENSDART00000143295 |
hnmt
|
histamine N-methyltransferase |
| chr2_+_42005217 | 0.23 |
ENSDART00000143562
|
gbp2
|
guanylate binding protein 2 |
| chr4_+_10721795 | 0.23 |
ENSDART00000136000
ENSDART00000067253 |
stab2
|
stabilin 2 |
| chr5_+_30596822 | 0.23 |
ENSDART00000188375
|
hinfp
|
histone H4 transcription factor |
| chr12_-_31422433 | 0.22 |
ENSDART00000186075
ENSDART00000153172 ENSDART00000066256 |
vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr22_-_18546241 | 0.22 |
ENSDART00000105404
ENSDART00000105405 |
cirbpb
|
cold inducible RNA binding protein b |
| chr21_-_44731865 | 0.20 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr19_-_6988837 | 0.20 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr20_+_13969414 | 0.19 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr13_+_6219551 | 0.19 |
ENSDART00000025910
|
apmap
|
adipocyte plasma membrane associated protein |
| chr1_+_59146298 | 0.18 |
ENSDART00000191885
ENSDART00000152747 |
gpr108
|
G protein-coupled receptor 108 |
| chr7_+_53156810 | 0.18 |
ENSDART00000189816
|
cdh29
|
cadherin 29 |
| chr2_+_46589798 | 0.18 |
ENSDART00000128457
ENSDART00000145347 |
ephb1
|
EPH receptor B1 |
| chr10_-_44411032 | 0.18 |
ENSDART00000111509
|
CABZ01072096.1
|
|
| chr3_+_19621034 | 0.16 |
ENSDART00000025358
|
itgb3a
|
integrin beta 3a |
| chr13_-_40754499 | 0.16 |
ENSDART00000111641
ENSDART00000159255 |
morn4
|
MORN repeat containing 4 |
| chr14_+_15430991 | 0.15 |
ENSDART00000158221
|
si:dkey-203a12.5
|
si:dkey-203a12.5 |
| chr6_-_30859656 | 0.15 |
ENSDART00000156235
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr21_+_261490 | 0.14 |
ENSDART00000177919
|
jak2a
|
Janus kinase 2a |
| chr16_-_30885838 | 0.14 |
ENSDART00000131356
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr2_+_42005475 | 0.14 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr17_-_27200634 | 0.14 |
ENSDART00000185332
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr6_-_31326398 | 0.13 |
ENSDART00000190749
ENSDART00000192789 |
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr19_-_34995629 | 0.13 |
ENSDART00000141704
|
si:rp71-45k5.2
|
si:rp71-45k5.2 |
| chr4_-_2219705 | 0.13 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr5_+_69878693 | 0.12 |
ENSDART00000073671
|
ythdc1
|
YTH domain containing 1 |
| chr10_-_45379831 | 0.12 |
ENSDART00000186205
|
CABZ01117348.1
|
|
| chr8_-_14121634 | 0.12 |
ENSDART00000184946
|
bgna
|
biglycan a |
| chr22_+_15979430 | 0.11 |
ENSDART00000189703
ENSDART00000192674 |
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr19_+_13994563 | 0.11 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr20_-_28642061 | 0.11 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr23_-_27152866 | 0.11 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr10_-_35542071 | 0.11 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr2_-_30784502 | 0.11 |
ENSDART00000056735
|
rgs20
|
regulator of G protein signaling 20 |
| chr12_+_30367371 | 0.11 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr15_-_38129845 | 0.11 |
ENSDART00000057095
|
si:dkey-24p1.1
|
si:dkey-24p1.1 |
| chr25_+_32474031 | 0.10 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr13_+_31583034 | 0.10 |
ENSDART00000111763
|
six6a
|
SIX homeobox 6a |
| chr1_-_470812 | 0.10 |
ENSDART00000192527
|
zgc:92518
|
zgc:92518 |
| chr16_-_43041324 | 0.09 |
ENSDART00000155445
ENSDART00000156836 ENSDART00000154945 |
si:dkey-7j14.6
|
si:dkey-7j14.6 |
| chr12_+_30367079 | 0.09 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr21_+_13389499 | 0.09 |
ENSDART00000151268
|
zgc:113162
|
zgc:113162 |
| chr3_+_34121156 | 0.08 |
ENSDART00000174929
|
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr18_+_30567945 | 0.08 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr6_+_11850359 | 0.08 |
ENSDART00000109552
ENSDART00000188139 ENSDART00000181499 ENSDART00000178269 |
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr6_+_28796773 | 0.08 |
ENSDART00000168237
ENSDART00000163541 |
tp63
|
tumor protein p63 |
| chr16_-_22930925 | 0.08 |
ENSDART00000133819
|
si:dkey-246i14.3
|
si:dkey-246i14.3 |
| chr23_+_4414343 | 0.07 |
ENSDART00000081821
|
wnt7ab
|
wingless-type MMTV integration site family, member 7Ab |
| chr19_+_49721 | 0.07 |
ENSDART00000160489
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr4_-_11077872 | 0.07 |
ENSDART00000150566
|
BX784029.4
|
|
| chr5_+_40224938 | 0.07 |
ENSDART00000142897
|
si:dkey-193c22.2
|
si:dkey-193c22.2 |
| chr5_-_25620594 | 0.07 |
ENSDART00000189346
|
cyp1d1
|
cytochrome P450, family 1, subfamily D, polypeptide 1 |
| chr25_-_14637660 | 0.06 |
ENSDART00000143666
|
nav2b
|
neuron navigator 2b |
| chr9_-_46399611 | 0.05 |
ENSDART00000164914
ENSDART00000145931 |
lct
si:dkey-79p17.3
|
lactase si:dkey-79p17.3 |
| chr23_-_22130778 | 0.05 |
ENSDART00000079212
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr23_-_33038423 | 0.05 |
ENSDART00000180539
|
plxna2
|
plexin A2 |
| chr15_+_7992906 | 0.05 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr23_+_4414164 | 0.04 |
ENSDART00000192762
|
wnt7ab
|
wingless-type MMTV integration site family, member 7Ab |
| chr10_+_43188678 | 0.04 |
ENSDART00000012522
|
vcanb
|
versican b |
| chr4_+_27130412 | 0.04 |
ENSDART00000145083
|
brd1a
|
bromodomain containing 1a |
| chr10_+_5645887 | 0.04 |
ENSDART00000171426
|
pdzph1
|
PDZ and pleckstrin homology domains 1 |
| chr9_+_56194410 | 0.04 |
ENSDART00000168530
|
LO018176.1
|
|
| chr3_-_8285123 | 0.04 |
ENSDART00000158699
ENSDART00000138588 |
trim35-9
|
tripartite motif containing 35-9 |
| chr18_-_16395668 | 0.04 |
ENSDART00000186004
|
mgat4c
|
mgat4 family, member C |
| chr20_+_35484070 | 0.03 |
ENSDART00000026234
ENSDART00000141675 |
mep1a.2
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 2 |
| chr11_+_14295011 | 0.03 |
ENSDART00000060226
|
si:ch211-262i1.4
|
si:ch211-262i1.4 |
| chr5_+_36439405 | 0.03 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr4_+_28997595 | 0.03 |
ENSDART00000133357
|
si:dkey-13e3.1
|
si:dkey-13e3.1 |
| chr6_-_11768198 | 0.03 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr1_+_57235896 | 0.03 |
ENSDART00000152621
|
si:dkey-27j5.7
|
si:dkey-27j5.7 |
| chr23_+_34005792 | 0.03 |
ENSDART00000132668
|
si:ch211-207e14.4
|
si:ch211-207e14.4 |
| chr4_+_77160027 | 0.03 |
ENSDART00000174060
ENSDART00000174364 |
znf1009
|
zinc finger protein 1009 |
| chr2_-_38337122 | 0.03 |
ENSDART00000076523
ENSDART00000187473 |
slc7a8b
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8b |
| chr8_+_1065458 | 0.03 |
ENSDART00000081432
|
sprb
|
sepiapterin reductase b |
| chr21_-_21465111 | 0.02 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr4_-_77506362 | 0.02 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr21_+_11560153 | 0.02 |
ENSDART00000065842
|
cd8a
|
CD8a molecule |
| chr24_-_21587335 | 0.02 |
ENSDART00000091528
|
gpr12
|
G protein-coupled receptor 12 |
| chr15_-_35212462 | 0.02 |
ENSDART00000043960
|
agfg1a
|
ArfGAP with FG repeats 1a |
| chr4_-_72101234 | 0.02 |
ENSDART00000167048
|
slco1f1
|
solute carrier organic anion transporter family, member 1F1 |
| chr10_-_40490647 | 0.02 |
ENSDART00000143660
|
taar20x
|
trace amine associated receptor 20x |
| chr4_+_63253425 | 0.01 |
ENSDART00000193510
|
si:ch211-258f1.3
|
si:ch211-258f1.3 |
| chr4_-_17741513 | 0.01 |
ENSDART00000141680
|
mybpc1
|
myosin binding protein C, slow type |
| chr4_-_5856200 | 0.01 |
ENSDART00000121936
|
slc5a8
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr1_+_59090583 | 0.01 |
ENSDART00000150658
|
mfap4
|
microfibril associated protein 4 |
| chr4_+_58016732 | 0.01 |
ENSDART00000165777
|
CT027609.1
|
|
| chr13_-_38039871 | 0.01 |
ENSDART00000140645
|
CR456624.1
|
|
| chr22_-_24858042 | 0.01 |
ENSDART00000137998
ENSDART00000078216 ENSDART00000138378 |
vtg7
|
vitellogenin 7 |
| chrM_+_9735 | 0.01 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr22_+_31207799 | 0.01 |
ENSDART00000133267
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr22_+_38762693 | 0.00 |
ENSDART00000015016
ENSDART00000150187 |
alpi.1
|
alkaline phosphatase, intestinal, tandem duplicate 1 |
| chr25_+_32473433 | 0.00 |
ENSDART00000152326
|
sqor
|
sulfide quinone oxidoreductase |
| chr20_+_27194833 | 0.00 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr22_-_3261879 | 0.00 |
ENSDART00000159643
|
gpr35.1
|
G protein-coupled receptor 35, tandem duplicate 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gli1+gli2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.2 | 0.7 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.1 | 0.7 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.6 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 0.6 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.7 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.1 | 0.8 | GO:2000095 | cerebrospinal fluid circulation(GO:0090660) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.3 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 0.3 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 1.0 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.8 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.3 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.4 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.5 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.4 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.0 | 0.7 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.4 | GO:0060260 | regulation of transcription initiation from RNA polymerase II promoter(GO:0060260) regulation of DNA-templated transcription, initiation(GO:2000142) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.8 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.5 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.4 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.5 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.3 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.7 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 0.6 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.4 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.3 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.4 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.5 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.5 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 1.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.1 | 0.8 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.7 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 1.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 1.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |