Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
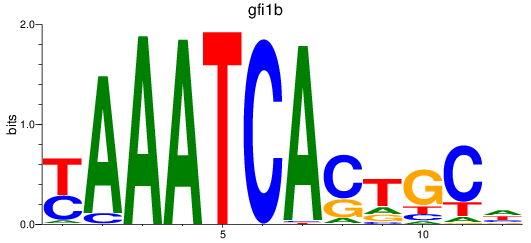
Results for gfi1b
Z-value: 0.81
Transcription factors associated with gfi1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gfi1b
|
ENSDARG00000079947 | growth factor independent 1B transcription repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gfi1b | dr11_v1_chr21_-_17290941_17290941 | -0.30 | 2.2e-01 | Click! |
Activity profile of gfi1b motif
Sorted Z-values of gfi1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_15937610 | 1.96 |
ENSDART00000061134
ENSDART00000154505 |
itpr2
|
inositol 1,4,5-trisphosphate receptor, type 2 |
| chr21_+_7605803 | 1.66 |
ENSDART00000121813
|
wdr41
|
WD repeat domain 41 |
| chr11_+_31121340 | 1.50 |
ENSDART00000185172
|
stx10
|
syntaxin 10 |
| chr7_+_69187585 | 1.48 |
ENSDART00000160499
ENSDART00000166258 |
marveld3
|
MARVEL domain containing 3 |
| chr3_-_36440705 | 1.35 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr3_-_54607166 | 1.27 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr10_-_35257458 | 1.26 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr25_-_17918536 | 1.21 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr5_-_45958838 | 1.20 |
ENSDART00000135072
|
poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr2_+_9821757 | 1.17 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr7_+_35191220 | 1.12 |
ENSDART00000110552
|
zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr24_-_23716097 | 1.11 |
ENSDART00000084954
ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr13_+_15816573 | 1.09 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr13_-_40282770 | 1.06 |
ENSDART00000140875
|
zgc:123010
|
zgc:123010 |
| chr21_+_31253048 | 1.06 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr18_+_27511976 | 1.03 |
ENSDART00000132017
ENSDART00000140781 |
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr8_+_10862353 | 1.01 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr11_-_35171768 | 1.00 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr6_-_12912606 | 1.00 |
ENSDART00000164640
|
ical1
|
islet cell autoantigen 1-like |
| chr3_+_51684963 | 0.97 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr11_-_35171162 | 0.96 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr16_+_26612401 | 0.96 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr20_+_6535176 | 0.96 |
ENSDART00000054652
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr20_+_6535438 | 0.95 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr6_+_37655078 | 0.90 |
ENSDART00000122199
ENSDART00000065127 |
cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr12_+_38563073 | 0.90 |
ENSDART00000009172
|
ttyh2
|
tweety family member 2 |
| chr22_+_34784075 | 0.90 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr11_+_13424116 | 0.90 |
ENSDART00000125563
|
homer3b
|
homer scaffolding protein 3b |
| chr13_-_42724645 | 0.88 |
ENSDART00000046066
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr5_-_24029228 | 0.87 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr9_-_19489264 | 0.86 |
ENSDART00000122894
|
wdr4
|
WD repeat domain 4 |
| chr25_+_418932 | 0.81 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr2_+_9822319 | 0.80 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr11_+_13423776 | 0.80 |
ENSDART00000102553
|
homer3b
|
homer scaffolding protein 3b |
| chr25_-_17918810 | 0.79 |
ENSDART00000023959
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr1_-_59240975 | 0.78 |
ENSDART00000166170
|
mvb12a
|
multivesicular body subunit 12A |
| chr24_+_35387517 | 0.77 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr3_-_37148594 | 0.77 |
ENSDART00000140855
|
mlx
|
MLX, MAX dimerization protein |
| chr16_-_44945224 | 0.76 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr13_+_22712406 | 0.74 |
ENSDART00000132847
|
si:ch211-134m17.9
|
si:ch211-134m17.9 |
| chr16_+_10329701 | 0.74 |
ENSDART00000172845
|
mdc1
|
mediator of DNA damage checkpoint 1 |
| chr16_+_39159752 | 0.73 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr7_-_33684632 | 0.73 |
ENSDART00000130553
|
tle3b
|
transducin-like enhancer of split 3b |
| chr18_-_22735002 | 0.72 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr14_-_33277743 | 0.72 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr2_+_37836821 | 0.71 |
ENSDART00000143203
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr14_-_33278084 | 0.70 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr16_-_41667101 | 0.70 |
ENSDART00000084528
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr12_-_3778848 | 0.70 |
ENSDART00000152128
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr11_+_18612166 | 0.69 |
ENSDART00000162694
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr8_-_15182232 | 0.68 |
ENSDART00000138855
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr11_-_2297832 | 0.68 |
ENSDART00000158266
|
znf740a
|
zinc finger protein 740a |
| chr19_+_20201254 | 0.68 |
ENSDART00000010140
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr8_+_11425048 | 0.66 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr18_-_38270430 | 0.65 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr18_-_38270077 | 0.65 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr18_-_35459996 | 0.65 |
ENSDART00000141023
|
itpkcb
|
inositol-trisphosphate 3-kinase Cb |
| chr5_+_60928576 | 0.65 |
ENSDART00000131041
|
doc2b
|
double C2-like domains, beta |
| chr7_+_13609457 | 0.64 |
ENSDART00000172857
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr18_-_35459605 | 0.63 |
ENSDART00000135691
|
itpkcb
|
inositol-trisphosphate 3-kinase Cb |
| chr24_-_25096199 | 0.62 |
ENSDART00000185076
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr19_-_425145 | 0.61 |
ENSDART00000164905
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr4_-_20235904 | 0.60 |
ENSDART00000146621
ENSDART00000193655 |
stk38l
|
serine/threonine kinase 38 like |
| chr6_+_50393047 | 0.60 |
ENSDART00000055502
ENSDART00000055511 |
ergic3
|
ERGIC and golgi 3 |
| chr5_+_44064764 | 0.60 |
ENSDART00000143843
|
si:dkey-84j12.1
|
si:dkey-84j12.1 |
| chr16_-_6849754 | 0.60 |
ENSDART00000149206
ENSDART00000149778 |
mbpb
|
myelin basic protein b |
| chr24_+_21684189 | 0.60 |
ENSDART00000014696
|
pdx1
|
pancreatic and duodenal homeobox 1 |
| chr1_+_35495368 | 0.60 |
ENSDART00000053806
|
gab1
|
GRB2-associated binding protein 1 |
| chr19_+_20201593 | 0.60 |
ENSDART00000163026
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr3_-_42086577 | 0.58 |
ENSDART00000083111
ENSDART00000187312 |
ttyh3a
|
tweety family member 3a |
| chr2_-_15040345 | 0.58 |
ENSDART00000109657
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr20_-_45772306 | 0.57 |
ENSDART00000062092
|
trmt6
|
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr16_-_31824525 | 0.57 |
ENSDART00000058737
|
cdc42l
|
cell division cycle 42, like |
| chr12_+_19199735 | 0.55 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr7_+_39054478 | 0.54 |
ENSDART00000173825
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr19_-_12193622 | 0.53 |
ENSDART00000041960
|
ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr22_+_22437561 | 0.53 |
ENSDART00000089622
|
kif14
|
kinesin family member 14 |
| chr23_-_18415872 | 0.51 |
ENSDART00000135430
|
fam120c
|
family with sequence similarity 120C |
| chr13_+_15657911 | 0.51 |
ENSDART00000134972
ENSDART00000138991 ENSDART00000133342 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr13_+_13681681 | 0.49 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr11_-_6452444 | 0.49 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr4_+_6833735 | 0.48 |
ENSDART00000136355
|
dock4b
|
dedicator of cytokinesis 4b |
| chr9_+_54984900 | 0.48 |
ENSDART00000191622
|
mospd2
|
motile sperm domain containing 2 |
| chr4_-_75057322 | 0.47 |
ENSDART00000157935
|
large1
|
LARGE xylosyl- and glucuronyltransferase 1 |
| chr6_-_24103666 | 0.47 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr9_+_54984537 | 0.47 |
ENSDART00000029528
|
mospd2
|
motile sperm domain containing 2 |
| chr4_+_6833583 | 0.46 |
ENSDART00000165179
ENSDART00000186134 ENSDART00000174507 |
dock4b
|
dedicator of cytokinesis 4b |
| chr19_+_40322912 | 0.45 |
ENSDART00000146893
|
cdk6
|
cyclin-dependent kinase 6 |
| chr20_+_4157815 | 0.44 |
ENSDART00000113132
|
gnpat
|
glyceronephosphate O-acyltransferase |
| chr9_+_44722205 | 0.43 |
ENSDART00000086176
ENSDART00000145271 ENSDART00000132696 |
nckap1
|
NCK-associated protein 1 |
| chr7_+_22801465 | 0.42 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr7_-_66133786 | 0.42 |
ENSDART00000154961
|
btbd10b
|
BTB (POZ) domain containing 10b |
| chr2_+_34967022 | 0.41 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr5_+_43965078 | 0.41 |
ENSDART00000113502
ENSDART00000187143 |
TMEM8B
|
si:dkey-84j12.1 |
| chr6_-_3924543 | 0.41 |
ENSDART00000170584
|
tlk1b
|
tousled-like kinase 1b |
| chr23_-_29505463 | 0.41 |
ENSDART00000050915
|
kif1b
|
kinesin family member 1B |
| chr6_-_3924723 | 0.40 |
ENSDART00000171804
|
tlk1b
|
tousled-like kinase 1b |
| chr9_+_48007081 | 0.40 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr8_-_51954562 | 0.39 |
ENSDART00000132527
ENSDART00000057315 |
cep78
|
centrosomal protein 78 |
| chr7_+_30201611 | 0.38 |
ENSDART00000075588
|
wdr76
|
WD repeat domain 76 |
| chr14_+_49007501 | 0.38 |
ENSDART00000128508
|
zdhhc5b
|
zinc finger, DHHC-type containing 5b |
| chr17_+_9308425 | 0.38 |
ENSDART00000188283
ENSDART00000183311 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr7_+_30202104 | 0.37 |
ENSDART00000173525
|
wdr76
|
WD repeat domain 76 |
| chr5_+_57773222 | 0.36 |
ENSDART00000135344
|
ppp2r1ba
|
protein phosphatase 2, regulatory subunit A, beta a |
| chr12_+_21299338 | 0.35 |
ENSDART00000074540
ENSDART00000133188 |
ca10a
|
carbonic anhydrase Xa |
| chr11_+_35171406 | 0.35 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr23_-_3721444 | 0.35 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr19_-_31707892 | 0.35 |
ENSDART00000088427
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr2_+_31838442 | 0.34 |
ENSDART00000066789
|
stard3nl
|
STARD3 N-terminal like |
| chr7_+_7019911 | 0.34 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr2_+_34967210 | 0.34 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr23_-_29505645 | 0.34 |
ENSDART00000146458
|
kif1b
|
kinesin family member 1B |
| chr19_-_9503473 | 0.32 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr4_+_13615731 | 0.32 |
ENSDART00000141395
ENSDART00000144310 |
ccdc87
|
coiled-coil domain containing 87 |
| chr4_-_14624481 | 0.31 |
ENSDART00000137847
|
plxnb2a
|
plexin b2a |
| chr17_+_49127003 | 0.31 |
ENSDART00000157360
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr7_+_9981757 | 0.31 |
ENSDART00000113429
ENSDART00000173233 |
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr17_-_37195163 | 0.31 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr17_+_24687338 | 0.30 |
ENSDART00000135794
|
selenon
|
selenoprotein N |
| chr20_-_43917647 | 0.29 |
ENSDART00000026213
|
mark3b
|
MAP/microtubule affinity-regulating kinase 3b |
| chr8_-_16515127 | 0.29 |
ENSDART00000146469
ENSDART00000132681 |
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr11_-_6877973 | 0.29 |
ENSDART00000160271
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr14_-_47391084 | 0.28 |
ENSDART00000159608
|
fstl5
|
follistatin-like 5 |
| chr13_-_24396199 | 0.26 |
ENSDART00000181093
|
tbp
|
TATA box binding protein |
| chr17_+_34805897 | 0.26 |
ENSDART00000137090
ENSDART00000077626 |
id2a
|
inhibitor of DNA binding 2a |
| chr11_-_24532988 | 0.25 |
ENSDART00000067078
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr14_-_31151148 | 0.24 |
ENSDART00000026569
|
gpc4
|
glypican 4 |
| chr9_+_19489304 | 0.24 |
ENSDART00000151920
|
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr3_-_30941362 | 0.22 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chr6_+_45494227 | 0.22 |
ENSDART00000159863
|
cntn4
|
contactin 4 |
| chr9_+_6255973 | 0.22 |
ENSDART00000078525
|
uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr7_-_72208248 | 0.21 |
ENSDART00000108916
|
zmp:0000001168
|
zmp:0000001168 |
| chr12_+_4845448 | 0.21 |
ENSDART00000160666
|
ghrhr2
|
growth hormone releasing hormone receptor 2 |
| chr9_+_6255682 | 0.21 |
ENSDART00000149827
|
uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr23_-_10137322 | 0.21 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr14_+_35023923 | 0.20 |
ENSDART00000172171
|
ebf3a
|
early B cell factor 3a |
| chr7_+_28960518 | 0.20 |
ENSDART00000182903
ENSDART00000188250 ENSDART00000191830 |
dok4
|
docking protein 4 |
| chr12_-_26022663 | 0.19 |
ENSDART00000166769
|
bmpr1ab
|
bone morphogenetic protein receptor, type IAb |
| chr14_-_9713549 | 0.18 |
ENSDART00000193356
ENSDART00000166739 |
si:zfos-2326c3.2
|
si:zfos-2326c3.2 |
| chr3_+_23691847 | 0.16 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr3_+_36972586 | 0.16 |
ENSDART00000102784
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr20_-_29864390 | 0.16 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr14_-_45512807 | 0.14 |
ENSDART00000173172
|
si:ch211-114c17.1
|
si:ch211-114c17.1 |
| chr4_-_27897160 | 0.13 |
ENSDART00000066924
ENSDART00000066925 ENSDART00000193020 |
tbc1d22a
|
TBC1 domain family, member 22a |
| chr8_-_1266181 | 0.13 |
ENSDART00000148654
ENSDART00000149924 |
cdc14b
|
cell division cycle 14B |
| chr19_+_4892281 | 0.12 |
ENSDART00000150969
|
cdk12
|
cyclin-dependent kinase 12 |
| chr21_-_11327830 | 0.12 |
ENSDART00000122331
|
rtkn2b
|
rhotekin 2b |
| chr5_-_37871526 | 0.11 |
ENSDART00000136450
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr7_+_39360797 | 0.11 |
ENSDART00000173481
|
acp2
|
acid phosphatase 2, lysosomal |
| chr20_+_13783040 | 0.10 |
ENSDART00000115329
ENSDART00000152497 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr16_-_17560822 | 0.09 |
ENSDART00000089230
|
casp2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr19_-_28130658 | 0.08 |
ENSDART00000079114
|
irx1b
|
iroquois homeobox 1b |
| chr5_+_36439405 | 0.08 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr3_+_19299309 | 0.07 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr3_+_36972298 | 0.07 |
ENSDART00000150917
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr21_+_26733529 | 0.07 |
ENSDART00000168379
|
pcxa
|
pyruvate carboxylase a |
| chr13_-_9119867 | 0.07 |
ENSDART00000137255
|
si:dkey-112g5.15
|
si:dkey-112g5.15 |
| chr9_+_3055566 | 0.06 |
ENSDART00000189906
ENSDART00000175891 ENSDART00000093021 |
ppp1r9ala
|
protein phosphatase 1 regulatory subunit 9A-like A |
| chr9_-_1965727 | 0.05 |
ENSDART00000082354
|
hoxd9a
|
homeobox D9a |
| chr23_+_25232711 | 0.05 |
ENSDART00000128510
|
erbb3b
|
erb-b2 receptor tyrosine kinase 3b |
| chr12_+_8474868 | 0.05 |
ENSDART00000062858
|
adoa
|
2-aminoethanethiol (cysteamine) dioxygenase a |
| chr17_-_37395460 | 0.05 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr1_+_18863060 | 0.05 |
ENSDART00000139241
|
rnf38
|
ring finger protein 38 |
| chr8_+_37111643 | 0.04 |
ENSDART00000061336
|
ren
|
renin |
| chr19_+_29798064 | 0.04 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr20_+_34596205 | 0.04 |
ENSDART00000138338
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr4_-_42242844 | 0.04 |
ENSDART00000163476
|
si:ch211-129p6.2
|
si:ch211-129p6.2 |
| chr17_-_33552363 | 0.03 |
ENSDART00000154400
|
tmem121aa
|
transmembrane protein 121Aa |
| chr16_+_10422836 | 0.02 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr10_-_34867401 | 0.02 |
ENSDART00000145545
|
dclk1a
|
doublecortin-like kinase 1a |
| chr23_+_25232411 | 0.01 |
ENSDART00000138974
|
erbb3b
|
erb-b2 receptor tyrosine kinase 3b |
| chr19_-_31584444 | 0.01 |
ENSDART00000052183
|
zgc:111986
|
zgc:111986 |
| chr5_-_23715861 | 0.01 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr14_-_6943934 | 0.01 |
ENSDART00000126279
|
clk4a
|
CDC-like kinase 4a |
| chr5_-_58832332 | 0.01 |
ENSDART00000161230
|
arhgef12b
|
Rho guanine nucleotide exchange factor (GEF) 12b |
| chr9_+_13638329 | 0.01 |
ENSDART00000143432
|
als2a
|
amyotrophic lateral sclerosis 2a (juvenile) |
| chr16_-_45235947 | 0.00 |
ENSDART00000164436
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr16_-_54978981 | 0.00 |
ENSDART00000154023
|
wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr1_+_1805294 | 0.00 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gfi1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.3 | 0.9 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.3 | 1.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.3 | 0.8 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.3 | 1.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 0.7 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.2 | 1.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 0.7 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.2 | 0.9 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.2 | 0.9 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 0.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.4 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.7 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 1.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.3 | GO:2000402 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.0 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.4 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.3 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.6 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 2.3 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 1.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.6 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.2 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.1 | 0.7 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 2.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.4 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.0 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.8 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.7 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.2 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 1.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 1.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.9 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 0.1 | GO:0048939 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 0.6 | GO:0071910 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.5 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 1.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.6 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 1.5 | GO:0006821 | chloride transport(GO:0006821) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 1.4 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 2.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 2.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.0 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 2.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 1.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.3 | 1.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.3 | 1.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 1.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 2.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 0.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.7 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.6 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.0 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.7 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.4 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.7 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.9 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.5 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 2.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 2.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 1.3 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |