Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
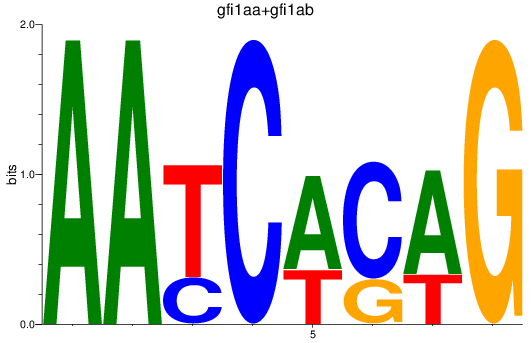
Results for gfi1aa+gfi1ab
Z-value: 0.81
Transcription factors associated with gfi1aa+gfi1ab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gfi1aa
|
ENSDARG00000020746 | growth factor independent 1A transcription repressor a |
|
gfi1ab
|
ENSDARG00000044457 | growth factor independent 1A transcription repressor b |
|
gfi1ab
|
ENSDARG00000114140 | growth factor independent 1A transcription repressor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gfi1aa | dr11_v1_chr2_+_10766744_10766744 | -0.64 | 4.0e-03 | Click! |
| gfi1ab | dr11_v1_chr6_-_29007493_29007493 | -0.57 | 1.4e-02 | Click! |
Activity profile of gfi1aa+gfi1ab motif
Sorted Z-values of gfi1aa+gfi1ab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_35252761 | 1.84 |
ENSDART00000051278
|
tnpo1
|
transportin 1 |
| chr11_-_10456387 | 1.78 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr14_-_49110280 | 1.70 |
ENSDART00000023654
|
sfxn1
|
sideroflexin 1 |
| chr11_-_10456553 | 1.69 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr6_-_53143667 | 1.63 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr20_+_26940178 | 1.60 |
ENSDART00000190888
|
cdca4
|
cell division cycle associated 4 |
| chr2_-_9915814 | 1.54 |
ENSDART00000091644
ENSDART00000177556 |
abi1b
|
abl-interactor 1b |
| chr2_+_23007675 | 1.52 |
ENSDART00000163649
|
mknk2a
|
MAP kinase interacting serine/threonine kinase 2a |
| chr7_-_26603743 | 1.50 |
ENSDART00000099003
|
plscr3b
|
phospholipid scramblase 3b |
| chr5_+_57328535 | 1.43 |
ENSDART00000149646
|
slc31a1
|
solute carrier family 31 (copper transporter), member 1 |
| chr10_-_15849027 | 1.41 |
ENSDART00000184682
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr3_+_54168007 | 1.38 |
ENSDART00000109894
|
olfm2a
|
olfactomedin 2a |
| chr3_-_36440705 | 1.37 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr21_+_13233377 | 1.33 |
ENSDART00000142569
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr9_+_54984900 | 1.33 |
ENSDART00000191622
|
mospd2
|
motile sperm domain containing 2 |
| chr20_+_27749133 | 1.32 |
ENSDART00000089013
|
vrtn
|
vertebrae development associated |
| chr7_-_60351876 | 1.31 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr13_+_15816573 | 1.30 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr20_+_26939742 | 1.29 |
ENSDART00000138369
ENSDART00000062061 ENSDART00000152992 |
cdca4
|
cell division cycle associated 4 |
| chr6_-_39518489 | 1.25 |
ENSDART00000185446
|
atf1
|
activating transcription factor 1 |
| chr16_-_51151993 | 1.25 |
ENSDART00000156255
|
ago1
|
argonaute RISC catalytic component 1 |
| chr7_-_60351537 | 1.24 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr18_+_35130416 | 1.24 |
ENSDART00000151595
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr25_+_36292465 | 1.21 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr5_-_54714525 | 1.20 |
ENSDART00000150138
ENSDART00000150070 |
ccnb1
|
cyclin B1 |
| chr5_-_54714789 | 1.20 |
ENSDART00000063357
|
ccnb1
|
cyclin B1 |
| chr14_-_7245971 | 1.20 |
ENSDART00000108796
|
stox2b
|
storkhead box 2b |
| chr24_+_39137001 | 1.19 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr25_-_17918536 | 1.18 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr6_-_53326421 | 1.17 |
ENSDART00000191740
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr2_+_9821757 | 1.17 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr25_+_7532811 | 1.16 |
ENSDART00000161593
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr10_+_41939963 | 1.14 |
ENSDART00000126248
|
tmem120b
|
transmembrane protein 120B |
| chr21_-_9466201 | 1.14 |
ENSDART00000167211
|
ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr16_+_39159752 | 1.14 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr7_+_69187585 | 1.14 |
ENSDART00000160499
ENSDART00000166258 |
marveld3
|
MARVEL domain containing 3 |
| chr16_-_54187374 | 1.11 |
ENSDART00000186989
ENSDART00000190113 |
LO017872.1
|
|
| chr4_+_9279784 | 1.10 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr8_+_16758304 | 1.09 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr24_+_12835935 | 1.09 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr3_-_54607166 | 1.09 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr6_-_49537646 | 1.09 |
ENSDART00000180438
|
FO704848.1
|
|
| chr24_-_26622423 | 1.08 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr13_-_18691041 | 1.07 |
ENSDART00000057867
|
sfxn3
|
sideroflexin 3 |
| chr6_+_36821621 | 1.07 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr21_+_38033226 | 1.06 |
ENSDART00000085728
|
klf8
|
Kruppel-like factor 8 |
| chr11_+_11120532 | 1.05 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr4_-_12477224 | 1.03 |
ENSDART00000027756
ENSDART00000182706 ENSDART00000127150 |
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr24_+_19518303 | 1.02 |
ENSDART00000027022
ENSDART00000056080 |
sulf1
|
sulfatase 1 |
| chr13_-_45022527 | 1.02 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr20_-_25645150 | 1.01 |
ENSDART00000063137
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr15_+_30126971 | 1.00 |
ENSDART00000100214
|
nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr9_-_34500197 | 0.99 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr18_+_22793465 | 0.99 |
ENSDART00000149685
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr10_+_38663917 | 0.98 |
ENSDART00000170182
|
acat1
|
acetyl-CoA acetyltransferase 1 |
| chr4_-_1720648 | 0.97 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr1_-_40519340 | 0.97 |
ENSDART00000114659
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr24_+_19518570 | 0.97 |
ENSDART00000056081
|
sulf1
|
sulfatase 1 |
| chr16_+_45337663 | 0.97 |
ENSDART00000147343
ENSDART00000185731 |
dnase2
|
deoxyribonuclease II, lysosomal |
| chr16_-_34285106 | 0.97 |
ENSDART00000044235
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr17_+_14965570 | 0.96 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr17_-_22573311 | 0.96 |
ENSDART00000141523
ENSDART00000140022 ENSDART00000079390 ENSDART00000188644 |
exo1
|
exonuclease 1 |
| chr6_+_40563848 | 0.96 |
ENSDART00000154766
|
si:ch73-15b2.5
|
si:ch73-15b2.5 |
| chr20_-_31427390 | 0.95 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| chr4_-_20177868 | 0.95 |
ENSDART00000003621
|
sinup
|
siaz-interacting nuclear protein |
| chr16_+_30117798 | 0.94 |
ENSDART00000135723
ENSDART00000000198 |
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr8_-_7308373 | 0.94 |
ENSDART00000132009
ENSDART00000145345 |
grip2a
|
glutamate receptor interacting protein 2a |
| chr8_+_10869183 | 0.94 |
ENSDART00000188111
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr22_-_10541712 | 0.93 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr5_-_25572151 | 0.92 |
ENSDART00000144995
|
si:dkey-229d2.4
|
si:dkey-229d2.4 |
| chr14_-_33278084 | 0.92 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr11_-_30508843 | 0.91 |
ENSDART00000101667
ENSDART00000179930 |
map4k3a
|
mitogen-activated protein kinase kinase kinase kinase 3a |
| chr14_-_33277743 | 0.91 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr12_-_28794957 | 0.90 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr16_+_43077909 | 0.90 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr13_+_22712406 | 0.89 |
ENSDART00000132847
|
si:ch211-134m17.9
|
si:ch211-134m17.9 |
| chr13_-_45022301 | 0.89 |
ENSDART00000183589
ENSDART00000125633 ENSDART00000074787 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr17_-_31483469 | 0.87 |
ENSDART00000062907
ENSDART00000061547 |
ltk
|
leukocyte receptor tyrosine kinase |
| chr3_+_26244353 | 0.87 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr11_+_13424116 | 0.86 |
ENSDART00000125563
|
homer3b
|
homer scaffolding protein 3b |
| chr4_-_20235904 | 0.86 |
ENSDART00000146621
ENSDART00000193655 |
stk38l
|
serine/threonine kinase 38 like |
| chr23_+_1216215 | 0.86 |
ENSDART00000165957
|
utrn
|
utrophin |
| chr25_-_17918810 | 0.86 |
ENSDART00000023959
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr6_+_40523370 | 0.86 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr12_+_16087077 | 0.86 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr12_+_10706772 | 0.86 |
ENSDART00000158227
|
top2a
|
DNA topoisomerase II alpha |
| chr3_+_32661765 | 0.85 |
ENSDART00000165562
|
si:dkey-16l2.16
|
si:dkey-16l2.16 |
| chr20_-_13625588 | 0.85 |
ENSDART00000078893
|
sytl3
|
synaptotagmin-like 3 |
| chr13_-_48431766 | 0.85 |
ENSDART00000159688
ENSDART00000171765 |
fbxo11a
|
F-box protein 11a |
| chr13_+_11829072 | 0.83 |
ENSDART00000079356
ENSDART00000170160 |
sufu
|
suppressor of fused homolog (Drosophila) |
| chr11_+_19370717 | 0.83 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr17_-_8638713 | 0.82 |
ENSDART00000148971
|
ctbp2a
|
C-terminal binding protein 2a |
| chr15_-_17025212 | 0.82 |
ENSDART00000014465
|
hip1
|
huntingtin interacting protein 1 |
| chr5_-_31773208 | 0.82 |
ENSDART00000137556
ENSDART00000122066 |
fam102ab
|
family with sequence similarity 102, member Ab |
| chr13_+_47710434 | 0.81 |
ENSDART00000188724
|
TMEM87B
|
transmembrane protein 87B |
| chr20_+_23625387 | 0.81 |
ENSDART00000147945
ENSDART00000150497 |
palld
|
palladin, cytoskeletal associated protein |
| chr23_-_26228077 | 0.81 |
ENSDART00000162423
|
BX927204.1
|
|
| chr9_+_33340311 | 0.79 |
ENSDART00000140064
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr6_+_38626926 | 0.78 |
ENSDART00000190339
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr18_+_22217489 | 0.78 |
ENSDART00000165464
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr22_-_10541372 | 0.77 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr18_-_38270430 | 0.77 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr21_-_929448 | 0.76 |
ENSDART00000133976
|
txnl1
|
thioredoxin-like 1 |
| chr12_-_48188928 | 0.76 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr15_-_9294929 | 0.76 |
ENSDART00000155717
|
si:ch211-261a10.5
|
si:ch211-261a10.5 |
| chr5_-_54712159 | 0.76 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr19_+_791538 | 0.75 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr15_-_31265375 | 0.75 |
ENSDART00000086592
|
vezf1b
|
vascular endothelial zinc finger 1b |
| chr2_+_9822319 | 0.75 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr3_+_26294161 | 0.75 |
ENSDART00000157648
ENSDART00000172337 |
rhot1a
|
ras homolog family member T1a |
| chr13_-_24263682 | 0.74 |
ENSDART00000176800
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr20_-_51814080 | 0.74 |
ENSDART00000041476
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr19_+_37120491 | 0.74 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr5_-_45958838 | 0.74 |
ENSDART00000135072
|
poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr3_-_33113879 | 0.74 |
ENSDART00000044677
|
rarab
|
retinoic acid receptor, alpha b |
| chr22_+_30137374 | 0.74 |
ENSDART00000187808
|
add3a
|
adducin 3 (gamma) a |
| chr23_+_9268083 | 0.73 |
ENSDART00000055054
|
acss2
|
acyl-CoA synthetase short chain family member 2 |
| chr20_-_28842524 | 0.73 |
ENSDART00000046035
ENSDART00000139843 ENSDART00000129858 ENSDART00000137425 ENSDART00000135720 |
max
|
myc associated factor X |
| chr8_+_3649036 | 0.73 |
ENSDART00000033152
|
rab35b
|
RAB35, member RAS oncogene family b |
| chr24_-_27256673 | 0.73 |
ENSDART00000181182
|
zmynd11
|
zinc finger, MYND-type containing 11 |
| chr3_+_29082267 | 0.73 |
ENSDART00000145615
|
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr6_+_27090800 | 0.73 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr19_-_27448606 | 0.73 |
ENSDART00000079251
|
vars2
|
valyl-tRNA synthetase 2, mitochondrial (putative) |
| chr3_-_34561624 | 0.72 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr5_-_69180227 | 0.72 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr17_-_37795865 | 0.72 |
ENSDART00000025853
|
zfp36l1a
|
zinc finger protein 36, C3H type-like 1a |
| chr11_+_14104417 | 0.72 |
ENSDART00000059752
ENSDART00000186575 |
med16
|
mediator complex subunit 16 |
| chr23_-_31645760 | 0.72 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr22_-_506522 | 0.71 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr15_-_17024779 | 0.70 |
ENSDART00000154719
|
hip1
|
huntingtin interacting protein 1 |
| chr9_+_22634073 | 0.70 |
ENSDART00000181822
|
etv5a
|
ets variant 5a |
| chr12_-_33817804 | 0.70 |
ENSDART00000191385
|
twnk
|
twinkle mtDNA helicase |
| chr2_-_21786826 | 0.69 |
ENSDART00000016208
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr13_-_42724645 | 0.69 |
ENSDART00000046066
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr9_+_28232522 | 0.69 |
ENSDART00000031761
|
fzd5
|
frizzled class receptor 5 |
| chr22_-_11493236 | 0.69 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr5_-_69934558 | 0.68 |
ENSDART00000124954
|
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr23_+_27778670 | 0.68 |
ENSDART00000053863
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr7_+_56735195 | 0.68 |
ENSDART00000082830
|
KIAA0895L
|
KIAA0895 like |
| chr6_-_12912606 | 0.68 |
ENSDART00000164640
|
ical1
|
islet cell autoantigen 1-like |
| chr9_+_54984537 | 0.67 |
ENSDART00000029528
|
mospd2
|
motile sperm domain containing 2 |
| chr8_-_16674584 | 0.67 |
ENSDART00000100727
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr3_-_26244256 | 0.67 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr8_-_36618073 | 0.67 |
ENSDART00000047912
|
gpkow
|
G patch domain and KOW motifs |
| chr24_-_21973365 | 0.67 |
ENSDART00000081204
ENSDART00000030592 |
acot9.1
|
acyl-CoA thioesterase 9, tandem duplicate 1 |
| chr8_+_54081819 | 0.67 |
ENSDART00000005857
ENSDART00000161795 |
prickle2a
|
prickle homolog 2a |
| chr6_+_50393047 | 0.66 |
ENSDART00000055502
ENSDART00000055511 |
ergic3
|
ERGIC and golgi 3 |
| chr12_-_8958353 | 0.65 |
ENSDART00000041728
|
cyp26a1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr18_+_20047374 | 0.65 |
ENSDART00000146957
|
uacaa
|
uveal autoantigen with coiled-coil domains and ankyrin repeats a |
| chr1_-_46650022 | 0.65 |
ENSDART00000148759
ENSDART00000053222 |
rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr15_-_17169935 | 0.64 |
ENSDART00000110111
|
cul5a
|
cullin 5a |
| chr20_+_25645459 | 0.63 |
ENSDART00000143555
ENSDART00000036350 |
aasdh
|
aminoadipate-semialdehyde dehydrogenase |
| chr19_-_8798178 | 0.63 |
ENSDART00000188232
|
cers2a
|
ceramide synthase 2a |
| chr5_+_31959954 | 0.63 |
ENSDART00000142826
|
myo1hb
|
myosin IHb |
| chr9_-_22057658 | 0.63 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr17_+_30591287 | 0.62 |
ENSDART00000154243
|
si:dkey-190l8.2
|
si:dkey-190l8.2 |
| chr2_+_36620011 | 0.62 |
ENSDART00000177428
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr15_+_17100697 | 0.62 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr18_+_27571448 | 0.62 |
ENSDART00000147886
|
cd82b
|
CD82 molecule b |
| chr3_-_5228137 | 0.62 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr22_+_22437561 | 0.62 |
ENSDART00000089622
|
kif14
|
kinesin family member 14 |
| chr24_+_39277043 | 0.61 |
ENSDART00000165458
|
MPRIP
|
si:ch73-103b11.2 |
| chr24_-_21973163 | 0.61 |
ENSDART00000131406
|
acot9.1
|
acyl-CoA thioesterase 9, tandem duplicate 1 |
| chr16_-_31284922 | 0.61 |
ENSDART00000142638
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr7_+_66884570 | 0.61 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr18_-_20466061 | 0.61 |
ENSDART00000060311
|
paqr5a
|
progestin and adipoQ receptor family member Va |
| chr11_-_43473824 | 0.61 |
ENSDART00000179561
|
tmem63bb
|
transmembrane protein 63Bb |
| chr1_+_10003193 | 0.60 |
ENSDART00000162675
|
trim2b
|
tripartite motif containing 2b |
| chr3_-_58165254 | 0.60 |
ENSDART00000093031
|
snu13a
|
SNU13 homolog, small nuclear ribonucleoprotein a (U4/U6.U5) |
| chr22_+_25088999 | 0.59 |
ENSDART00000158225
|
rrbp1b
|
ribosome binding protein 1b |
| chr15_-_41310340 | 0.59 |
ENSDART00000192112
|
si:dkey-75b17.1
|
si:dkey-75b17.1 |
| chr16_+_26601364 | 0.58 |
ENSDART00000087537
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr6_-_14040136 | 0.58 |
ENSDART00000065361
ENSDART00000179765 |
etv5b
|
ets variant 5b |
| chr8_+_16738282 | 0.58 |
ENSDART00000134265
ENSDART00000100698 |
ercc8
|
excision repair cross-complementation group 8 |
| chr11_+_25693395 | 0.58 |
ENSDART00000110224
|
mon1bb
|
MON1 secretory trafficking family member Bb |
| chr20_-_25902141 | 0.58 |
ENSDART00000142611
ENSDART00000024821 |
elmsan1a
|
ELM2 and Myb/SANT-like domain containing 1a |
| chr17_+_19630068 | 0.57 |
ENSDART00000182619
|
rgs7a
|
regulator of G protein signaling 7a |
| chr21_-_27619701 | 0.57 |
ENSDART00000133441
ENSDART00000180100 |
pcnxl3
|
pecanex-like 3 (Drosophila) |
| chr8_-_25566347 | 0.57 |
ENSDART00000138289
ENSDART00000078022 |
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr7_+_34236238 | 0.57 |
ENSDART00000052474
|
tipin
|
timeless interacting protein |
| chr12_+_32368574 | 0.57 |
ENSDART00000086389
|
ANKFN1
|
si:ch211-277e21.2 |
| chr19_+_27448625 | 0.57 |
ENSDART00000052352
|
ppp1r11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr13_+_43650632 | 0.57 |
ENSDART00000141024
|
zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr10_+_7719796 | 0.57 |
ENSDART00000191795
|
ggcx
|
gamma-glutamyl carboxylase |
| chr6_-_46053300 | 0.57 |
ENSDART00000169722
|
ca16b
|
carbonic anhydrase XVI b |
| chr16_+_26439518 | 0.57 |
ENSDART00000041787
|
trim35-28
|
tripartite motif containing 35-28 |
| chr2_+_31308587 | 0.57 |
ENSDART00000027090
|
clul1
|
clusterin-like 1 (retinal) |
| chr18_+_16722129 | 0.56 |
ENSDART00000146163
|
eif4g2b
|
eukaryotic translation initiation factor 4, gamma 2b |
| chr7_-_48396193 | 0.56 |
ENSDART00000083555
|
sin3ab
|
SIN3 transcription regulator family member Ab |
| chr2_-_40889465 | 0.56 |
ENSDART00000192631
ENSDART00000180824 |
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr23_+_19594608 | 0.56 |
ENSDART00000134865
|
slmapb
|
sarcolemma associated protein b |
| chr11_+_14333441 | 0.56 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr16_-_42770064 | 0.55 |
ENSDART00000112879
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr6_+_59642695 | 0.55 |
ENSDART00000166373
ENSDART00000161030 |
R3HDM2
|
R3H domain containing 2 |
| chr7_+_17063761 | 0.54 |
ENSDART00000182880
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr3_-_1400309 | 0.54 |
ENSDART00000159893
|
wbp11
|
WW domain binding protein 11 |
| chr18_-_25905574 | 0.54 |
ENSDART00000143899
ENSDART00000163369 |
sema4ba
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ba |
| chr18_-_38270077 | 0.54 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr18_+_44703343 | 0.54 |
ENSDART00000131510
|
b3gnt2l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2, like |
| chr23_-_32157865 | 0.54 |
ENSDART00000000876
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gfi1aa+gfi1ab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.4 | 1.1 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.4 | 3.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.3 | 1.7 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.3 | 3.6 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.3 | 1.4 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.3 | 1.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 2.0 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.2 | 0.7 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.2 | 0.7 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 1.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 0.8 | GO:1900181 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.2 | 1.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 2.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 2.5 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.1 | 1.0 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 1.6 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.4 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.1 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 1.0 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 1.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.7 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.5 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.9 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 0.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.7 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 1.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.5 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 0.4 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.1 | 1.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.8 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.5 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 1.3 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.6 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.1 | 0.6 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.8 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.6 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.3 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.4 | GO:0071674 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of lamellipodium organization(GO:1902745) |
| 0.1 | 0.6 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.3 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.7 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 0.3 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 1.5 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 1.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.9 | GO:0001840 | neural plate development(GO:0001840) |
| 0.1 | 0.7 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.7 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 0.7 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 1.0 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.3 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 1.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.9 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.2 | GO:0039529 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.1 | 0.3 | GO:2000252 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.4 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 2.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 1.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 0.2 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 1.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.2 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.3 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.1 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.4 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.3 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.1 | 0.5 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.1 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.3 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.5 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 1.4 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.2 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.4 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 1.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 1.1 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.3 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.5 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.0 | 0.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.7 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 1.5 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.4 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.5 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 1.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 1.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.7 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 0.3 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.0 | 0.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 2.4 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.0 | 1.3 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 2.4 | GO:0051170 | nuclear import(GO:0051170) |
| 0.0 | 0.2 | GO:0048909 | anterior lateral line nerve development(GO:0048909) |
| 0.0 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 2.5 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 2.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 1.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) vasodilation(GO:0042311) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 4.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.6 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.2 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.0 | 1.0 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 1.0 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.0 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 1.2 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.7 | GO:0060389 | pathway-restricted SMAD protein phosphorylation(GO:0060389) |
| 0.0 | 0.4 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.3 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.4 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.5 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.4 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.2 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.0 | 0.6 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.5 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.0 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.3 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.3 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 1.1 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.8 | GO:0030335 | positive regulation of cell migration(GO:0030335) |
| 0.0 | 0.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.2 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.6 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.0 | GO:0035268 | protein mannosylation(GO:0035268) mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.0 | 0.1 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.6 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.3 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) regulation of chromatin assembly(GO:0010847) |
| 0.0 | 1.3 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.1 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.2 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 0.2 | GO:0014855 | striated muscle cell proliferation(GO:0014855) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.4 | 1.5 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.3 | 1.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 0.7 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.2 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 0.8 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 0.6 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.7 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.4 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 3.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 1.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.9 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 2.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 1.2 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 2.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 1.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 2.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.3 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.5 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 0.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 1.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.3 | 1.0 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.3 | 1.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.3 | 1.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.7 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 0.7 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 0.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.7 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.7 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 0.6 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 1.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.4 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.1 | 2.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 1.8 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.0 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 1.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 1.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 3.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.5 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.7 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.7 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 1.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.9 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.0 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 2.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 1.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.5 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 1.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 1.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.9 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.5 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.4 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.4 | GO:0003916 | DNA topoisomerase activity(GO:0003916) DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 3.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.6 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.2 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.7 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.0 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 4.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 1.4 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 1.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.9 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 3.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.6 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 1.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.9 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 3.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.3 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.1 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |