Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
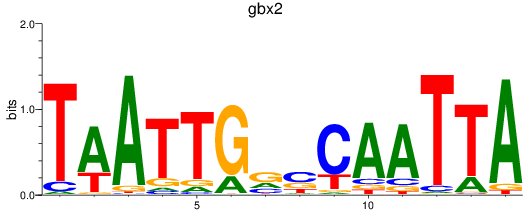
Results for gbx2
Z-value: 0.81
Transcription factors associated with gbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gbx2
|
ENSDARG00000002933 | gastrulation brain homeobox 2 |
|
gbx2
|
ENSDARG00000116377 | gastrulation brain homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gbx2 | dr11_v1_chr6_+_16031189_16031189 | -0.31 | 2.1e-01 | Click! |
Activity profile of gbx2 motif
Sorted Z-values of gbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_6452444 | 4.46 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr1_-_18811517 | 2.43 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr7_-_33684632 | 2.38 |
ENSDART00000130553
|
tle3b
|
transducin-like enhancer of split 3b |
| chr7_+_46019780 | 2.35 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr7_-_51727760 | 2.29 |
ENSDART00000174180
|
hdac8
|
histone deacetylase 8 |
| chr17_+_24318753 | 2.21 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr12_-_35386910 | 2.16 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr8_+_23826985 | 2.10 |
ENSDART00000187430
|
mapk14a
|
mitogen-activated protein kinase 14a |
| chr6_+_3717613 | 1.66 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr7_+_59677273 | 1.60 |
ENSDART00000039535
ENSDART00000132044 |
trmt44
|
tRNA methyltransferase 44 homolog |
| chr20_-_49889111 | 1.58 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr22_+_980290 | 1.54 |
ENSDART00000065377
|
def6b
|
differentially expressed in FDCP 6b homolog (mouse) |
| chr19_-_25149034 | 1.43 |
ENSDART00000148432
ENSDART00000175266 |
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr11_-_12158412 | 1.43 |
ENSDART00000147670
|
npepps
|
aminopeptidase puromycin sensitive |
| chr19_-_25149598 | 1.35 |
ENSDART00000162917
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr7_-_58098814 | 1.23 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr8_+_21229718 | 1.20 |
ENSDART00000100222
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr18_-_16937008 | 1.15 |
ENSDART00000100117
ENSDART00000022640 ENSDART00000136541 |
znf143b
|
zinc finger protein 143b |
| chr24_-_33801668 | 1.10 |
ENSDART00000079202
|
abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr10_+_43797130 | 1.07 |
ENSDART00000027242
|
nr2f1b
|
nuclear receptor subfamily 2, group F, member 1b |
| chr10_-_14929630 | 1.06 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
| chr9_+_23003208 | 1.06 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr13_-_33654931 | 0.93 |
ENSDART00000020350
|
snx5
|
sorting nexin 5 |
| chr14_+_35464994 | 0.85 |
ENSDART00000115307
|
si:ch211-203d1.3
|
si:ch211-203d1.3 |
| chr11_+_31323746 | 0.83 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr8_+_18010568 | 0.80 |
ENSDART00000121984
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr19_-_10330778 | 0.78 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr4_-_5019113 | 0.77 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr18_+_16750080 | 0.73 |
ENSDART00000136320
|
rnf141
|
ring finger protein 141 |
| chr11_+_2855430 | 0.73 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr15_-_10341048 | 0.72 |
ENSDART00000171013
|
tenm4
|
teneurin transmembrane protein 4 |
| chr22_-_14272699 | 0.70 |
ENSDART00000190121
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr9_+_711822 | 0.68 |
ENSDART00000136627
|
ybey
|
ybeY metallopeptidase |
| chr17_+_1496107 | 0.67 |
ENSDART00000187804
|
LO018430.1
|
|
| chr15_+_34934568 | 0.66 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr18_+_15644559 | 0.65 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr5_-_20123002 | 0.61 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr5_-_69923241 | 0.60 |
ENSDART00000187389
|
fktn
|
fukutin |
| chr5_-_25733745 | 0.58 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr7_+_20260172 | 0.58 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr17_+_49281597 | 0.56 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr11_+_16216909 | 0.56 |
ENSDART00000081035
ENSDART00000147190 |
slc25a26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr8_+_20140321 | 0.54 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr19_-_20430892 | 0.51 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr22_-_9861531 | 0.51 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr23_+_42254960 | 0.49 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr4_-_5018705 | 0.49 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr3_+_28581397 | 0.44 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr18_+_8833251 | 0.43 |
ENSDART00000143519
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr18_+_26895994 | 0.41 |
ENSDART00000098347
|
ch25hl1.2
|
cholesterol 25-hydroxylase like 1, tandem duplicate 2 |
| chr18_+_17537344 | 0.40 |
ENSDART00000025782
|
nup93
|
nucleoporin 93 |
| chr9_-_3934963 | 0.39 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr8_-_18010097 | 0.38 |
ENSDART00000122730
ENSDART00000133666 |
acot11b
|
acyl-CoA thioesterase 11b |
| chr20_-_18382708 | 0.36 |
ENSDART00000170864
ENSDART00000166762 ENSDART00000191333 |
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr24_-_32582378 | 0.36 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr14_-_14643190 | 0.35 |
ENSDART00000167119
|
znf185
|
zinc finger protein 185 with LIM domain |
| chr6_-_19270484 | 0.32 |
ENSDART00000186894
ENSDART00000188709 |
zgc:174863
|
zgc:174863 |
| chr8_+_23105117 | 0.31 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr22_+_2512154 | 0.30 |
ENSDART00000097363
|
zgc:173726
|
zgc:173726 |
| chr24_-_32582880 | 0.24 |
ENSDART00000186307
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr19_-_15855427 | 0.21 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr24_+_33802528 | 0.21 |
ENSDART00000136040
ENSDART00000147499 ENSDART00000182322 |
atg9b
|
autophagy related 9B |
| chr24_-_16904234 | 0.20 |
ENSDART00000192879
|
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr3_+_32118670 | 0.18 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr17_+_51499789 | 0.18 |
ENSDART00000187701
|
CABZ01067581.1
|
|
| chr23_-_333457 | 0.18 |
ENSDART00000114486
|
uhrf1bp1
|
UHRF1 binding protein 1 |
| chr10_-_39052264 | 0.17 |
ENSDART00000144036
|
igsf5a
|
immunoglobulin superfamily, member 5a |
| chr20_-_7128612 | 0.17 |
ENSDART00000146755
ENSDART00000036871 |
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr5_+_57773222 | 0.16 |
ENSDART00000135344
|
ppp2r1ba
|
protein phosphatase 2, regulatory subunit A, beta a |
| chr22_+_9862466 | 0.16 |
ENSDART00000146864
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr1_-_8000428 | 0.15 |
ENSDART00000133098
|
si:dkey-79f11.5
|
si:dkey-79f11.5 |
| chr7_-_3927813 | 0.14 |
ENSDART00000172991
ENSDART00000173392 |
si:dkey-88n24.12
|
si:dkey-88n24.12 |
| chr6_-_42369186 | 0.14 |
ENSDART00000039868
|
usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr15_-_5524271 | 0.11 |
ENSDART00000130297
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr10_-_11761927 | 0.10 |
ENSDART00000138748
|
adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr6_-_39218609 | 0.10 |
ENSDART00000133305
|
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr3_-_10739625 | 0.09 |
ENSDART00000156144
|
zgc:112965
|
zgc:112965 |
| chr12_+_22560067 | 0.07 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr12_-_20665865 | 0.07 |
ENSDART00000183922
|
gip
|
gastric inhibitory polypeptide |
| chr24_-_25364775 | 0.07 |
ENSDART00000151915
|
ptchd1
|
patched domain containing 1 |
| chr25_-_31702970 | 0.06 |
ENSDART00000153586
|
lamb4
|
laminin, beta 4 |
| chr9_+_55154414 | 0.05 |
ENSDART00000182924
|
ANOS1
|
anosmin 1 |
| chr11_-_422484 | 0.04 |
ENSDART00000122253
|
znf831
|
zinc finger protein 831 |
| chr19_-_27336092 | 0.04 |
ENSDART00000176885
|
gtf2h4
|
general transcription factor IIH, polypeptide 4 |
| chr15_-_12270857 | 0.04 |
ENSDART00000170093
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr24_-_26995164 | 0.04 |
ENSDART00000142864
|
stag1b
|
stromal antigen 1b |
| chr13_+_3252950 | 0.04 |
ENSDART00000020671
|
prph2b
|
peripherin 2b (retinal degeneration, slow) |
| chr8_+_31821396 | 0.03 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr1_+_40801696 | 0.03 |
ENSDART00000147497
|
cpz
|
carboxypeptidase Z |
| chr19_-_5265155 | 0.03 |
ENSDART00000145003
|
prf1.3
|
perforin 1.3 |
| chr5_+_30624183 | 0.02 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr8_+_44478294 | 0.02 |
ENSDART00000006898
|
si:ch73-211l2.3
|
si:ch73-211l2.3 |
| chr12_-_34713690 | 0.02 |
ENSDART00000180807
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr7_-_46019756 | 0.01 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr5_-_23843636 | 0.01 |
ENSDART00000193280
|
GBGT1 (1 of many)
|
si:ch211-135f11.5 |
| chr16_-_26296477 | 0.01 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr6_-_46790042 | 0.01 |
ENSDART00000156037
|
igfn1.4
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:1901741 | embryonic cleavage(GO:0040016) positive regulation of myoblast fusion(GO:1901741) |
| 0.3 | 2.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.2 | 1.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.2 | 1.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.7 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 2.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 1.3 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.1 | 0.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.6 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 2.2 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 1.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.1 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 2.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.1 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.0 | 0.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.6 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 2.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.3 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0019079 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 1.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 1.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 1.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 2.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.4 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 2.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.6 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.4 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 2.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.0 | 2.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 2.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |