Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
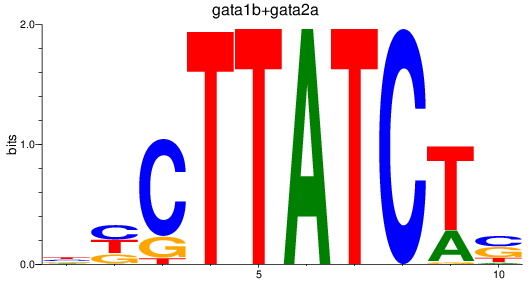
Results for gata1b+gata2a
Z-value: 1.02
Transcription factors associated with gata1b+gata2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gata1b
|
ENSDARG00000059130 | GATA binding protein 1b |
|
gata2a
|
ENSDARG00000059327 | GATA binding protein 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gata2a | dr11_v1_chr11_-_3865472_3865472 | 0.82 | 2.8e-05 | Click! |
| gata1b | dr11_v1_chr8_-_7474997_7474997 | 0.78 | 1.2e-04 | Click! |
Activity profile of gata1b+gata2a motif
Sorted Z-values of gata1b+gata2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_55105066 | 5.24 |
ENSDART00000110848
|
si:ch211-5k11.8
|
si:ch211-5k11.8 |
| chr13_+_2908764 | 3.36 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr3_+_55100045 | 3.13 |
ENSDART00000113098
|
hbaa1
|
hemoglobin, alpha adult 1 |
| chr10_-_29236860 | 2.76 |
ENSDART00000111620
|
ccdc83
|
coiled-coil domain containing 83 |
| chr23_-_23401305 | 2.69 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr7_+_35075847 | 2.59 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr23_+_7379728 | 2.58 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr16_-_24815091 | 2.50 |
ENSDART00000154269
ENSDART00000131025 |
kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr2_+_33051730 | 2.44 |
ENSDART00000177902
ENSDART00000187903 |
rnf220a
|
ring finger protein 220a |
| chr24_+_2843268 | 2.36 |
ENSDART00000170529
|
si:ch211-152c8.2
|
si:ch211-152c8.2 |
| chr23_-_35483163 | 2.34 |
ENSDART00000138660
ENSDART00000113643 ENSDART00000189269 |
fbxo25
|
F-box protein 25 |
| chr25_+_21829777 | 2.32 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr16_+_25126935 | 2.14 |
ENSDART00000058945
|
zgc:92590
|
zgc:92590 |
| chr20_+_15552657 | 1.97 |
ENSDART00000063912
|
jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr13_+_24679674 | 1.97 |
ENSDART00000033090
ENSDART00000139854 |
zgc:66426
|
zgc:66426 |
| chr2_+_33052360 | 1.92 |
ENSDART00000134651
|
rnf220a
|
ring finger protein 220a |
| chr12_+_47945664 | 1.75 |
ENSDART00000189824
ENSDART00000055743 ENSDART00000188936 |
CABZ01074994.1
|
|
| chr7_-_51102479 | 1.65 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr20_+_33981946 | 1.63 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr16_+_51180938 | 1.56 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr15_+_19838458 | 1.53 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr19_+_8606883 | 1.53 |
ENSDART00000054469
ENSDART00000185264 |
s100a10a
|
S100 calcium binding protein A10a |
| chr2_+_33052169 | 1.51 |
ENSDART00000180008
|
rnf220a
|
ring finger protein 220a |
| chr13_+_33304187 | 1.50 |
ENSDART00000075826
ENSDART00000145295 |
dcdc2b
|
doublecortin domain containing 2B |
| chr6_-_49078263 | 1.50 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr8_-_49431939 | 1.48 |
ENSDART00000011453
ENSDART00000088240 ENSDART00000114173 |
sypb
|
synaptophysin b |
| chr21_-_25756119 | 1.46 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr12_-_20350629 | 1.39 |
ENSDART00000066384
|
hbbe2
|
hemoglobin beta embryonic-2 |
| chr2_+_4207209 | 1.33 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr7_+_46368520 | 1.33 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr14_+_30568961 | 1.31 |
ENSDART00000184303
|
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr11_+_33818179 | 1.30 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr1_+_52563298 | 1.30 |
ENSDART00000142465
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr17_+_30894431 | 1.28 |
ENSDART00000127996
|
degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr1_-_45636362 | 1.26 |
ENSDART00000181757
|
sept3
|
septin 3 |
| chr6_+_58915889 | 1.24 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr1_-_8651718 | 1.23 |
ENSDART00000133319
|
actb1
|
actin, beta 1 |
| chr13_-_29406534 | 1.22 |
ENSDART00000100877
|
zgc:153142
|
zgc:153142 |
| chr8_-_39654669 | 1.22 |
ENSDART00000145677
|
si:dkey-63d15.12
|
si:dkey-63d15.12 |
| chr11_-_6265574 | 1.18 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr8_+_16004551 | 1.18 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr1_-_9109699 | 1.16 |
ENSDART00000147833
|
vap
|
vascular associated protein |
| chr14_-_25949713 | 1.14 |
ENSDART00000181455
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr19_+_4912817 | 1.12 |
ENSDART00000101658
ENSDART00000165082 |
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr11_-_28614608 | 1.12 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr14_+_17197132 | 1.10 |
ENSDART00000054598
|
rtn4rl2b
|
reticulon 4 receptor-like 2b |
| chr20_+_33875256 | 1.10 |
ENSDART00000002554
|
rxrgb
|
retinoid X receptor, gamma b |
| chr5_+_15203421 | 1.06 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr10_+_10801564 | 1.02 |
ENSDART00000027026
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr5_+_34622320 | 1.01 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr24_-_39858710 | 1.00 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr15_-_21877726 | 0.97 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr14_-_25949951 | 0.97 |
ENSDART00000141304
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr3_+_40809892 | 0.96 |
ENSDART00000190803
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr8_-_40327397 | 0.95 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr20_-_25533739 | 0.94 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr23_+_26079467 | 0.91 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr2_-_51794472 | 0.89 |
ENSDART00000186652
|
BX908782.3
|
|
| chr19_+_20778011 | 0.88 |
ENSDART00000024208
|
nutf2l
|
nuclear transport factor 2, like |
| chr13_-_4848889 | 0.87 |
ENSDART00000165259
|
mcu
|
mitochondrial calcium uniporter |
| chr16_-_36834505 | 0.86 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr8_+_31872992 | 0.84 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr2_+_21486529 | 0.82 |
ENSDART00000047468
|
inhbab
|
inhibin, beta Ab |
| chr10_-_17484762 | 0.81 |
ENSDART00000137905
ENSDART00000007961 |
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr2_+_38261748 | 0.80 |
ENSDART00000076478
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr25_-_7974494 | 0.80 |
ENSDART00000171446
|
hal
|
histidine ammonia-lyase |
| chr10_+_10801719 | 0.80 |
ENSDART00000193648
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr23_+_25708787 | 0.76 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr19_-_40985308 | 0.76 |
ENSDART00000132986
ENSDART00000142360 |
si:ch211-120e1.7
|
si:ch211-120e1.7 |
| chr3_-_12026741 | 0.74 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr14_-_28052474 | 0.73 |
ENSDART00000172948
ENSDART00000135337 |
TSC22D3 (1 of many)
zgc:64189
|
si:ch211-220e11.3 zgc:64189 |
| chr9_+_2522797 | 0.73 |
ENSDART00000186786
ENSDART00000147034 |
gpr155a
|
G protein-coupled receptor 155a |
| chr20_-_26420939 | 0.72 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr24_-_2843107 | 0.69 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr8_-_43574935 | 0.68 |
ENSDART00000051139
|
ncor2
|
nuclear receptor corepressor 2 |
| chr23_-_6920107 | 0.68 |
ENSDART00000149230
ENSDART00000182946 |
si:ch211-117c9.2
|
si:ch211-117c9.2 |
| chr13_+_844150 | 0.67 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr7_+_34290051 | 0.65 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr20_-_26421112 | 0.65 |
ENSDART00000183767
ENSDART00000182330 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr3_+_32492467 | 0.64 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr5_-_13645995 | 0.63 |
ENSDART00000099665
ENSDART00000166957 |
purba
|
purine-rich element binding protein Ba |
| chr9_+_29585943 | 0.63 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr19_+_7124337 | 0.61 |
ENSDART00000031380
|
tap2a
|
transporter associated with antigen processing, subunit type a |
| chr16_+_26449615 | 0.60 |
ENSDART00000039746
|
epb41b
|
erythrocyte membrane protein band 4.1b |
| chr21_-_11834452 | 0.60 |
ENSDART00000081652
|
rfesd
|
Rieske (Fe-S) domain containing |
| chr7_+_17120811 | 0.60 |
ENSDART00000147140
|
prmt3
|
protein arginine methyltransferase 3 |
| chr25_-_13728111 | 0.60 |
ENSDART00000169865
|
lcat
|
lecithin-cholesterol acyltransferase |
| chr9_-_45149695 | 0.59 |
ENSDART00000162033
|
CU467837.1
|
|
| chr6_-_40697585 | 0.59 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr21_+_27336550 | 0.56 |
ENSDART00000139634
|
dpp3
|
dipeptidyl-peptidase 3 |
| chr3_-_35602233 | 0.55 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr11_-_13126969 | 0.54 |
ENSDART00000169052
ENSDART00000190120 |
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr22_+_16535575 | 0.53 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr20_+_34870517 | 0.53 |
ENSDART00000191797
|
ankef1a
|
ankyrin repeat and EF-hand domain containing 1a |
| chr20_-_26066020 | 0.53 |
ENSDART00000078559
|
myct1a
|
myc target 1a |
| chr25_-_16826219 | 0.52 |
ENSDART00000191299
ENSDART00000188504 |
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr25_+_18564266 | 0.52 |
ENSDART00000172338
|
cav1
|
caveolin 1 |
| chr13_-_20381485 | 0.51 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr3_-_26204867 | 0.51 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr14_+_36738069 | 0.51 |
ENSDART00000105590
|
tdo2a
|
tryptophan 2,3-dioxygenase a |
| chr18_+_48428713 | 0.50 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr7_-_50604626 | 0.50 |
ENSDART00000073903
ENSDART00000174031 |
crtc3
|
CREB regulated transcription coactivator 3 |
| chr6_+_8176486 | 0.50 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr1_+_41854298 | 0.48 |
ENSDART00000192672
|
smox
|
spermine oxidase |
| chr22_-_36856405 | 0.48 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr6_-_49078702 | 0.47 |
ENSDART00000135185
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr24_-_10917243 | 0.47 |
ENSDART00000144372
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr24_+_29381946 | 0.47 |
ENSDART00000189551
|
ntng1a
|
netrin g1a |
| chr1_-_38816685 | 0.47 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr22_-_11626014 | 0.47 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr18_+_18405992 | 0.46 |
ENSDART00000080174
|
n4bp1
|
nedd4 binding protein 1 |
| chr10_+_26612321 | 0.46 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr18_-_16795262 | 0.45 |
ENSDART00000048722
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr5_-_42272517 | 0.45 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr11_-_1400507 | 0.45 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr5_-_66301142 | 0.44 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr23_-_33750307 | 0.44 |
ENSDART00000162772
|
bin2a
|
bridging integrator 2a |
| chr23_-_33750135 | 0.44 |
ENSDART00000187641
|
bin2a
|
bridging integrator 2a |
| chr4_-_23858900 | 0.44 |
ENSDART00000123199
|
usp6nl
|
USP6 N-terminal like |
| chr15_-_43164591 | 0.44 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr16_+_13424534 | 0.43 |
ENSDART00000114944
|
zgc:101640
|
zgc:101640 |
| chr9_+_343062 | 0.42 |
ENSDART00000164012
ENSDART00000164365 |
bpifcl
|
BPI fold containing family C, like |
| chr19_+_37509638 | 0.42 |
ENSDART00000139999
|
si:ch211-250g4.3
|
si:ch211-250g4.3 |
| chr8_+_4838114 | 0.42 |
ENSDART00000146667
|
es1
|
es1 protein |
| chr5_+_45921234 | 0.41 |
ENSDART00000134355
|
ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr8_-_14151051 | 0.41 |
ENSDART00000090427
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr10_-_43029001 | 0.41 |
ENSDART00000171494
|
ssbp2
|
single-stranded DNA binding protein 2 |
| chr14_-_41556720 | 0.41 |
ENSDART00000149244
|
itga6l
|
integrin, alpha 6, like |
| chr1_+_36674584 | 0.40 |
ENSDART00000186772
ENSDART00000192274 |
ednraa
|
endothelin receptor type Aa |
| chr8_+_16004154 | 0.40 |
ENSDART00000134787
ENSDART00000172510 ENSDART00000141173 |
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr21_-_11820379 | 0.38 |
ENSDART00000126640
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr15_+_26600611 | 0.38 |
ENSDART00000155352
|
slc47a3
|
solute carrier family 47 (multidrug and toxin extrusion), member 3 |
| chr10_+_40624702 | 0.37 |
ENSDART00000109129
|
zgc:172131
|
zgc:172131 |
| chr16_-_24668620 | 0.37 |
ENSDART00000012807
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr1_-_11291324 | 0.37 |
ENSDART00000091205
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr24_-_21258945 | 0.37 |
ENSDART00000111025
|
boc
|
BOC cell adhesion associated, oncogene regulated |
| chr14_+_35691889 | 0.36 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr8_+_20994433 | 0.36 |
ENSDART00000141736
|
si:dkeyp-82a1.8
|
si:dkeyp-82a1.8 |
| chr13_+_22480496 | 0.36 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr19_+_5146460 | 0.34 |
ENSDART00000150740
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr2_-_6482240 | 0.34 |
ENSDART00000132623
|
rgs13
|
regulator of G protein signaling 13 |
| chr24_+_29382109 | 0.34 |
ENSDART00000184620
ENSDART00000188414 ENSDART00000186132 ENSDART00000191489 |
ntng1a
|
netrin g1a |
| chr11_-_25418856 | 0.34 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr20_+_51813432 | 0.32 |
ENSDART00000127444
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr24_+_29449690 | 0.31 |
ENSDART00000105743
ENSDART00000193556 ENSDART00000145816 |
ntng1a
|
netrin g1a |
| chr25_+_35134393 | 0.31 |
ENSDART00000185379
|
CR762436.2
|
|
| chr23_-_24682244 | 0.31 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr1_-_17569793 | 0.30 |
ENSDART00000125125
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr5_-_16425781 | 0.30 |
ENSDART00000185624
ENSDART00000180617 |
slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr16_-_5612480 | 0.29 |
ENSDART00000157944
|
CR848788.1
|
|
| chr6_-_6673813 | 0.28 |
ENSDART00000150995
|
si:dkey-261j15.2
|
si:dkey-261j15.2 |
| chr5_-_51166025 | 0.28 |
ENSDART00000097471
|
card9
|
caspase recruitment domain family, member 9 |
| chr3_+_24275766 | 0.27 |
ENSDART00000055607
|
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr13_+_23677949 | 0.27 |
ENSDART00000144215
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr23_+_20408227 | 0.27 |
ENSDART00000134727
|
si:rp71-17i16.4
|
si:rp71-17i16.4 |
| chr2_+_47927026 | 0.26 |
ENSDART00000143023
|
ftr25
|
finTRIM family, member 25 |
| chr6_-_38418862 | 0.26 |
ENSDART00000104135
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr10_+_19525839 | 0.26 |
ENSDART00000162912
ENSDART00000158512 |
vsig8a
|
V-set and immunoglobulin domain containing 8a |
| chr7_+_44445595 | 0.26 |
ENSDART00000108766
|
cdh5
|
cadherin 5 |
| chr18_+_27732285 | 0.26 |
ENSDART00000141835
|
tspan18b
|
tetraspanin 18b |
| chr8_-_50287949 | 0.25 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr11_+_31285127 | 0.25 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr17_-_29736019 | 0.25 |
ENSDART00000183483
|
ush2a
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr15_-_568645 | 0.25 |
ENSDART00000156744
|
cbln18
|
cerebellin 18 |
| chr2_+_35240764 | 0.24 |
ENSDART00000015827
|
tnr
|
tenascin R (restrictin, janusin) |
| chr11_-_13126505 | 0.24 |
ENSDART00000158377
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr23_+_36115541 | 0.24 |
ENSDART00000130090
|
hoxc6a
|
homeobox C6a |
| chr13_+_10945337 | 0.24 |
ENSDART00000091845
|
abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr12_-_32421046 | 0.23 |
ENSDART00000075567
|
enpp7.1
|
ectonucleotide pyrophosphatase/phosphodiesterase 7, tandem duplicate 1 |
| chr8_+_24861264 | 0.22 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr18_+_17959681 | 0.22 |
ENSDART00000142700
|
znf423
|
zinc finger protein 423 |
| chr14_+_26546175 | 0.21 |
ENSDART00000105932
|
si:dkeyp-110e4.11
|
si:dkeyp-110e4.11 |
| chr17_+_8175998 | 0.20 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr6_-_13308813 | 0.20 |
ENSDART00000065372
|
kcnj3b
|
potassium inwardly-rectifying channel, subfamily J, member 3b |
| chr21_-_40880317 | 0.20 |
ENSDART00000100054
ENSDART00000137696 |
elnb
|
elastin b |
| chr20_-_27390258 | 0.19 |
ENSDART00000010584
|
prph2l
|
peripherin 2, like |
| chr23_+_42272588 | 0.19 |
ENSDART00000164907
|
CABZ01065131.1
|
|
| chr17_-_26867725 | 0.19 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr1_+_9954489 | 0.18 |
ENSDART00000005895
ENSDART00000183003 |
pdia2
|
protein disulfide isomerase family A, member 2 |
| chr5_+_21211135 | 0.18 |
ENSDART00000088492
|
bmp10
|
bone morphogenetic protein 10 |
| chr16_-_30564319 | 0.18 |
ENSDART00000145087
|
lmna
|
lamin A |
| chr17_+_39242437 | 0.18 |
ENSDART00000156138
ENSDART00000128863 |
zgc:174356
|
zgc:174356 |
| chr13_+_835390 | 0.17 |
ENSDART00000171329
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr10_+_21650828 | 0.17 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr18_+_45228691 | 0.17 |
ENSDART00000127953
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr8_+_23709280 | 0.17 |
ENSDART00000185842
|
ppardb
|
peroxisome proliferator-activated receptor delta b |
| chr16_-_12953739 | 0.17 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr24_-_25691020 | 0.17 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr6_-_609880 | 0.17 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr8_-_40555340 | 0.17 |
ENSDART00000163348
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr7_-_41851605 | 0.17 |
ENSDART00000142981
|
mylk3
|
myosin light chain kinase 3 |
| chr19_+_7567763 | 0.16 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr4_+_1530287 | 0.16 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr9_+_32978302 | 0.16 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr4_-_20118468 | 0.16 |
ENSDART00000078587
|
si:dkey-159a18.1
|
si:dkey-159a18.1 |
| chr12_-_36070473 | 0.15 |
ENSDART00000165720
|
gpr142
|
G protein-coupled receptor 142 |
| chr6_-_38419318 | 0.15 |
ENSDART00000138026
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr10_-_43718914 | 0.15 |
ENSDART00000189277
|
cetn3
|
centrin 3 |
| chr7_-_31938938 | 0.15 |
ENSDART00000132353
|
bdnf
|
brain-derived neurotrophic factor |
| chr18_+_33550667 | 0.14 |
ENSDART00000141246
ENSDART00000136070 |
si:dkey-47k20.1
|
si:dkey-47k20.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gata1b+gata2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.5 | 1.6 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.5 | 9.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.5 | 1.4 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.4 | 1.2 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.4 | 1.1 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.3 | 1.3 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.3 | 1.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.3 | 0.8 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.3 | 1.5 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.2 | 1.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.2 | 1.5 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.2 | 0.6 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.2 | 1.0 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.2 | 0.9 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.2 | 2.3 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 0.5 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.9 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.9 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 2.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.4 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 3.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.5 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 0.3 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.1 | 1.8 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 1.3 | GO:0034311 | diol metabolic process(GO:0034311) |
| 0.1 | 1.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.5 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.4 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 1.5 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 0.3 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 1.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 1.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.0 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 1.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 2.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.0 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 1.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.6 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.7 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.2 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 2.4 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0048901 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) anterior lateral line neuromast development(GO:0048901) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.3 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.0 | 1.0 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.4 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.6 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.4 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 1.6 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.0 | 0.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 8.5 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 1.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.2 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 1.5 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 1.2 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 0.9 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.9 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 1.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.9 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.1 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.3 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.0 | 1.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 2.0 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.5 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.5 | 2.0 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.4 | 1.6 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.3 | 2.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 1.3 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 1.2 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 1.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.0 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 1.0 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 2.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 0.5 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.2 | 0.8 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 1.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.6 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 0.5 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.6 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 2.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 2.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 1.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.8 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 1.1 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 4.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 3.9 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 5.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 2.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 2.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 2.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 2.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.5 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |