Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
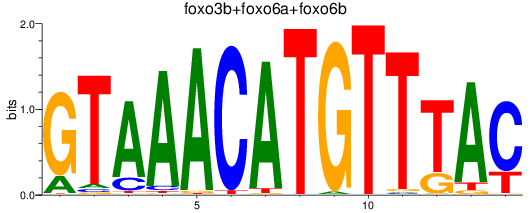
Results for foxo3b+foxo6a+foxo6b
Z-value: 0.46
Transcription factors associated with foxo3b+foxo6a+foxo6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxo6b
|
ENSDARG00000024619 | forkhead box O6 b |
|
foxo3b
|
ENSDARG00000042904 | forkhead box O3b |
|
foxo6a
|
ENSDARG00000100486 | forkhead box O6 a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxo6a | dr11_v1_chr16_-_7362806_7362806 | -0.88 | 1.4e-06 | Click! |
| foxo6b | dr11_v1_chr19_+_15571290_15571290 | -0.49 | 3.8e-02 | Click! |
| foxo3b | dr11_v1_chr20_-_32332736_32332736 | -0.42 | 8.5e-02 | Click! |
Activity profile of foxo3b+foxo6a+foxo6b motif
Sorted Z-values of foxo3b+foxo6a+foxo6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_28322986 | 1.80 |
ENSDART00000134710
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr22_+_25753972 | 1.39 |
ENSDART00000188417
|
AL929192.2
|
|
| chr15_+_38299563 | 1.27 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr15_+_38299385 | 1.20 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr5_+_25733774 | 1.05 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr9_+_27720428 | 1.03 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr9_+_426392 | 0.99 |
ENSDART00000172515
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr8_-_16464453 | 0.98 |
ENSDART00000098691
|
rnf11b
|
ring finger protein 11b |
| chr21_-_17037907 | 0.87 |
ENSDART00000101263
|
ube2g1b
|
ubiquitin-conjugating enzyme E2G 1b (UBC7 homolog, yeast) |
| chr12_-_48188928 | 0.84 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr9_-_27410597 | 0.79 |
ENSDART00000135652
ENSDART00000042297 |
kdelc1
|
KDEL (Lys-Asp-Glu-Leu) containing 1 |
| chr2_+_22409249 | 0.78 |
ENSDART00000182915
|
zgc:56628
|
zgc:56628 |
| chr23_-_36670369 | 0.78 |
ENSDART00000006881
|
zbtb39
|
zinc finger and BTB domain containing 39 |
| chr2_-_7131657 | 0.75 |
ENSDART00000175565
ENSDART00000092116 |
extl2
|
exostosin-like glycosyltransferase 2 |
| chr10_-_25570647 | 0.74 |
ENSDART00000134215
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr7_-_20241346 | 0.73 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr25_-_36261836 | 0.73 |
ENSDART00000179411
|
dus2
|
dihydrouridine synthase 2 |
| chr25_-_8160030 | 0.72 |
ENSDART00000067159
|
tph1a
|
tryptophan hydroxylase 1 (tryptophan 5-monooxygenase) a |
| chr5_-_12031174 | 0.69 |
ENSDART00000159896
|
castor1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr11_-_23697217 | 0.68 |
ENSDART00000124810
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr14_-_6402769 | 0.67 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr14_-_45551572 | 0.67 |
ENSDART00000111410
|
ganab
|
glucosidase, alpha; neutral AB |
| chr23_+_36308428 | 0.67 |
ENSDART00000134607
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr19_-_13286722 | 0.61 |
ENSDART00000168296
ENSDART00000158330 |
zfpm2b
|
zinc finger protein, FOG family member 2b |
| chr12_+_18970790 | 0.58 |
ENSDART00000153279
ENSDART00000153235 ENSDART00000078494 |
l3mbtl2
|
l(3)mbt-like 2 (Drosophila) |
| chr7_-_41881177 | 0.56 |
ENSDART00000174258
ENSDART00000018972 |
zgc:92818
|
zgc:92818 |
| chr23_-_912817 | 0.56 |
ENSDART00000192851
|
rhoac
|
ras homolog gene family, member Ac |
| chr2_-_37478418 | 0.54 |
ENSDART00000146103
|
dapk3
|
death-associated protein kinase 3 |
| chr20_-_24165509 | 0.53 |
ENSDART00000124919
|
map3k7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr17_+_51746830 | 0.52 |
ENSDART00000184230
|
odc1
|
ornithine decarboxylase 1 |
| chr5_+_29652513 | 0.52 |
ENSDART00000035400
|
tsc1a
|
TSC complex subunit 1a |
| chr22_-_21676364 | 0.51 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr11_+_14343445 | 0.51 |
ENSDART00000171347
ENSDART00000170547 |
si:ch211-262i1.6
|
si:ch211-262i1.6 |
| chr12_+_17603528 | 0.50 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr6_-_41138854 | 0.49 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr7_+_69449814 | 0.49 |
ENSDART00000109644
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr2_-_38080075 | 0.48 |
ENSDART00000056544
|
tox4a
|
TOX high mobility group box family member 4 a |
| chr12_-_28799642 | 0.48 |
ENSDART00000066303
|
mrpl10
|
mitochondrial ribosomal protein L10 |
| chr19_-_7441686 | 0.47 |
ENSDART00000168194
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr9_+_24088062 | 0.47 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr5_-_25733745 | 0.46 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr24_+_19414561 | 0.45 |
ENSDART00000193157
|
sulf1
|
sulfatase 1 |
| chr16_+_46410520 | 0.45 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr1_+_10018466 | 0.44 |
ENSDART00000113551
|
trim2b
|
tripartite motif containing 2b |
| chr24_+_32411753 | 0.44 |
ENSDART00000058530
|
neurod6a
|
neuronal differentiation 6a |
| chr5_+_28433215 | 0.42 |
ENSDART00000165072
ENSDART00000168005 |
nsmfb
|
NMDA receptor synaptonuclear signaling and neuronal migration factor b |
| chr23_-_12158685 | 0.42 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr12_+_46582168 | 0.42 |
ENSDART00000189402
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr19_-_30447611 | 0.42 |
ENSDART00000073705
ENSDART00000048977 ENSDART00000191237 |
abcf1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr13_-_13030851 | 0.42 |
ENSDART00000009499
|
nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr2_-_43850897 | 0.40 |
ENSDART00000005449
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr21_+_17301790 | 0.39 |
ENSDART00000145057
|
tsc1b
|
TSC complex subunit 1b |
| chr14_+_45350008 | 0.38 |
ENSDART00000025749
|
ttc9c
|
tetratricopeptide repeat domain 9C |
| chr16_+_53519048 | 0.38 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr15_-_19334448 | 0.36 |
ENSDART00000062576
|
thyn1
|
thymocyte nuclear protein 1 |
| chr24_+_19415124 | 0.36 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr7_-_50764714 | 0.36 |
ENSDART00000110283
|
iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr6_-_3998199 | 0.35 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr5_-_51998708 | 0.35 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr25_+_4855549 | 0.35 |
ENSDART00000163839
|
ap4e1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr19_-_7441948 | 0.34 |
ENSDART00000003544
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr3_+_9588705 | 0.34 |
ENSDART00000172543
ENSDART00000104875 |
trap1
|
TNF receptor-associated protein 1 |
| chr5_-_30145939 | 0.34 |
ENSDART00000086795
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr15_-_33304133 | 0.34 |
ENSDART00000186092
|
nbeab
|
neurobeachin b |
| chr8_+_18010978 | 0.34 |
ENSDART00000039887
ENSDART00000144532 |
ssbp3b
|
single stranded DNA binding protein 3b |
| chr16_+_26747766 | 0.33 |
ENSDART00000183257
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr20_+_33994580 | 0.33 |
ENSDART00000061729
|
si:dkey-97o5.1
|
si:dkey-97o5.1 |
| chr9_-_52386733 | 0.33 |
ENSDART00000171721
|
dap1b
|
death associated protein 1b |
| chr3_+_60828813 | 0.32 |
ENSDART00000128260
|
CABZ01087514.1
|
|
| chr16_-_25400257 | 0.32 |
ENSDART00000040756
|
zgc:136493
|
zgc:136493 |
| chr16_-_44878245 | 0.32 |
ENSDART00000154391
ENSDART00000154925 ENSDART00000154697 |
arhgap33
|
Rho GTPase activating protein 33 |
| chr21_-_3606539 | 0.32 |
ENSDART00000062418
|
dym
|
dymeclin |
| chr5_-_17876709 | 0.31 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr21_-_28640316 | 0.29 |
ENSDART00000128237
|
nrg2a
|
neuregulin 2a |
| chr9_-_12574473 | 0.29 |
ENSDART00000191372
ENSDART00000193667 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr22_+_2533514 | 0.28 |
ENSDART00000147967
|
si:ch73-92e7.4
|
si:ch73-92e7.4 |
| chr11_+_23933016 | 0.28 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr16_-_31351419 | 0.28 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr18_+_41527877 | 0.27 |
ENSDART00000146972
|
selenot1b
|
selenoprotein T, 1b |
| chr6_-_18531760 | 0.27 |
ENSDART00000167167
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr22_+_18319666 | 0.26 |
ENSDART00000033103
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr7_-_26049282 | 0.26 |
ENSDART00000136389
ENSDART00000101124 |
rnaseka
|
ribonuclease, RNase K a |
| chr7_+_40094081 | 0.25 |
ENSDART00000186054
|
si:ch73-174h16.4
|
si:ch73-174h16.4 |
| chr8_-_18010735 | 0.25 |
ENSDART00000125014
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr1_-_59567685 | 0.24 |
ENSDART00000159144
ENSDART00000182913 |
zmp:0000001082
|
zmp:0000001082 |
| chr6_+_55819038 | 0.24 |
ENSDART00000108786
|
si:ch211-81n22.1
|
si:ch211-81n22.1 |
| chr1_+_19708508 | 0.24 |
ENSDART00000054581
ENSDART00000131206 |
march1
|
membrane-associated ring finger (C3HC4) 1 |
| chr20_+_41801913 | 0.23 |
ENSDART00000139805
|
mcm9
|
minichromosome maintenance 9 homologous recombination repair factor |
| chr5_-_62940851 | 0.23 |
ENSDART00000137052
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr2_+_24317465 | 0.23 |
ENSDART00000139339
ENSDART00000136284 |
mrpl34
|
mitochondrial ribosomal protein L34 |
| chr3_+_52078798 | 0.22 |
ENSDART00000156882
|
si:dkey-88e12.3
|
si:dkey-88e12.3 |
| chr3_-_26341959 | 0.22 |
ENSDART00000169344
ENSDART00000142878 ENSDART00000087196 |
zgc:153240
|
zgc:153240 |
| chr7_+_30240791 | 0.22 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr1_-_45616242 | 0.22 |
ENSDART00000150066
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr7_-_36358303 | 0.22 |
ENSDART00000130028
|
fto
|
fat mass and obesity associated |
| chr25_-_35127617 | 0.22 |
ENSDART00000125128
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr24_-_25030173 | 0.21 |
ENSDART00000126395
ENSDART00000019350 |
nup58
|
nucleoporin 58 |
| chr7_+_32021669 | 0.21 |
ENSDART00000173976
|
mettl15
|
methyltransferase like 15 |
| chr18_+_21113216 | 0.21 |
ENSDART00000060191
|
kmt5b
|
lysine methyltransferase 5B |
| chr5_-_68795063 | 0.21 |
ENSDART00000016307
|
her1
|
hairy-related 1 |
| chr6_-_46560207 | 0.21 |
ENSDART00000187607
|
kcnb1
|
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr2_-_37797577 | 0.20 |
ENSDART00000110781
|
nfatc4
|
nuclear factor of activated T cells 4 |
| chr7_-_36358735 | 0.20 |
ENSDART00000188392
|
fto
|
fat mass and obesity associated |
| chr7_+_32021982 | 0.20 |
ENSDART00000173848
|
mettl15
|
methyltransferase like 15 |
| chr4_-_12781182 | 0.20 |
ENSDART00000058020
|
helb
|
helicase (DNA) B |
| chr22_-_10440688 | 0.20 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr23_+_42482137 | 0.19 |
ENSDART00000160199
|
cyp2aa3
|
cytochrome P450, family 2, subfamily AA, polypeptide 3 |
| chr17_+_24111392 | 0.19 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr25_+_28158352 | 0.18 |
ENSDART00000151854
|
cadps2
|
Ca++-dependent secretion activator 2 |
| chr7_-_57949117 | 0.17 |
ENSDART00000138803
|
ank2b
|
ankyrin 2b, neuronal |
| chr16_-_21692024 | 0.16 |
ENSDART00000123597
|
si:ch211-154o6.2
|
si:ch211-154o6.2 |
| chr5_-_31035198 | 0.16 |
ENSDART00000086534
|
cyb5d2
|
cytochrome b5 domain containing 2 |
| chr24_+_32472155 | 0.15 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr7_+_26006819 | 0.15 |
ENSDART00000112908
|
pelp1
|
proline, glutamate and leucine rich protein 1 |
| chr4_-_31666343 | 0.14 |
ENSDART00000190758
ENSDART00000192721 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr7_-_30174882 | 0.14 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr13_+_48358467 | 0.14 |
ENSDART00000171080
ENSDART00000162531 |
msh6
|
mutS homolog 6 (E. coli) |
| chr19_-_7690975 | 0.13 |
ENSDART00000151384
|
si:dkey-204a24.10
|
si:dkey-204a24.10 |
| chr12_-_4424424 | 0.12 |
ENSDART00000152314
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr5_+_56119975 | 0.12 |
ENSDART00000083137
|
mrm1
|
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr8_-_36125849 | 0.12 |
ENSDART00000159581
|
CT583723.1
|
|
| chr20_-_33583779 | 0.12 |
ENSDART00000097823
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr5_+_3118231 | 0.12 |
ENSDART00000179809
|
supt4h1
|
SPT4 homolog, DSIF elongation factor subunit |
| chr17_-_2834764 | 0.12 |
ENSDART00000024027
|
bdkrb1
|
bradykinin receptor B1 |
| chr9_-_3834686 | 0.11 |
ENSDART00000141492
|
myo3b
|
myosin IIIB |
| chr22_-_34995333 | 0.11 |
ENSDART00000110900
|
kcnip2
|
Kv channel interacting protein 2 |
| chr6_+_39279831 | 0.10 |
ENSDART00000155088
|
ankrd33ab
|
ankyrin repeat domain 33Ab |
| chr9_-_16877456 | 0.10 |
ENSDART00000161105
ENSDART00000160869 |
fbxl3a
|
F-box and leucine-rich repeat protein 3a |
| chr21_+_26539157 | 0.09 |
ENSDART00000021121
|
stx5al
|
syntaxin 5A, like |
| chr1_+_29515160 | 0.09 |
ENSDART00000152226
|
si:dkey-228l14.1
|
si:dkey-228l14.1 |
| chr10_+_38526496 | 0.09 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr20_+_22067337 | 0.09 |
ENSDART00000152636
|
clocka
|
clock circadian regulator a |
| chr11_-_7147540 | 0.09 |
ENSDART00000143942
|
bmp7a
|
bone morphogenetic protein 7a |
| chr10_-_36244864 | 0.08 |
ENSDART00000123748
|
or109-1
|
odorant receptor, family D, subfamily 109, member 1 |
| chr6_+_58549080 | 0.08 |
ENSDART00000180117
|
stmn3
|
stathmin-like 3 |
| chr8_+_47897734 | 0.08 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr16_-_29405009 | 0.08 |
ENSDART00000149289
|
tlr18
|
toll-like receptor 18 |
| chr1_+_56972616 | 0.08 |
ENSDART00000152812
|
si:ch211-1f22.11
|
si:ch211-1f22.11 |
| chr23_-_1017428 | 0.08 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr15_-_46736432 | 0.08 |
ENSDART00000124567
|
ilkap
|
integrin-linked kinase-associated serine/threonine phosphatase |
| chr3_-_34051363 | 0.07 |
ENSDART00000151768
|
ighv3-2
|
immunoglobulin heavy variable 3-2 |
| chr22_+_2524615 | 0.07 |
ENSDART00000134277
|
znf1003
|
zinc finger protein 1003 |
| chr4_+_38547203 | 0.07 |
ENSDART00000171349
|
si:ch211-209n20.1
|
si:ch211-209n20.1 |
| chr2_+_23130389 | 0.07 |
ENSDART00000142834
|
si:dkey-218h11.6
|
si:dkey-218h11.6 |
| chr19_-_19599372 | 0.07 |
ENSDART00000160002
ENSDART00000171664 |
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr1_-_11606903 | 0.07 |
ENSDART00000136093
|
si:dkey-26i13.6
|
si:dkey-26i13.6 |
| chr15_-_31357634 | 0.07 |
ENSDART00000127485
|
or111-2
|
odorant receptor, family D, subfamily 111, member 2 |
| chr7_-_32021853 | 0.07 |
ENSDART00000134521
|
kif18a
|
kinesin family member 18A |
| chr8_-_53044089 | 0.06 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr7_+_3597413 | 0.06 |
ENSDART00000064281
|
si:dkey-192d15.2
|
si:dkey-192d15.2 |
| chr22_-_23590069 | 0.06 |
ENSDART00000172067
|
f13b
|
coagulation factor XIII, B polypeptide |
| chr22_+_18319230 | 0.06 |
ENSDART00000184747
ENSDART00000184649 |
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr9_+_30211038 | 0.06 |
ENSDART00000190847
|
senp7a
|
SUMO1/sentrin specific peptidase 7a |
| chr11_-_21404044 | 0.06 |
ENSDART00000080116
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr5_+_29652198 | 0.06 |
ENSDART00000184083
|
tsc1a
|
TSC complex subunit 1a |
| chr21_-_43949208 | 0.06 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr1_-_11607062 | 0.06 |
ENSDART00000158253
|
si:dkey-26i13.6
|
si:dkey-26i13.6 |
| chr21_-_37973081 | 0.05 |
ENSDART00000136569
|
ripply1
|
ripply transcriptional repressor 1 |
| chr4_+_32772213 | 0.05 |
ENSDART00000137958
ENSDART00000125835 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr17_+_28670132 | 0.05 |
ENSDART00000076344
ENSDART00000164981 ENSDART00000182851 |
hectd1
|
HECT domain containing 1 |
| chr21_+_17541634 | 0.05 |
ENSDART00000191700
ENSDART00000163238 |
stom
|
stomatin |
| chr8_+_28695914 | 0.05 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr3_+_43460696 | 0.04 |
ENSDART00000164581
|
galr2b
|
galanin receptor 2b |
| chr10_-_7472323 | 0.04 |
ENSDART00000163702
ENSDART00000167054 ENSDART00000167706 |
nrg1
|
neuregulin 1 |
| chr23_+_17878969 | 0.04 |
ENSDART00000154934
ENSDART00000154140 ENSDART00000156331 |
si:ch73-390p7.2
|
si:ch73-390p7.2 |
| chr14_-_17306261 | 0.04 |
ENSDART00000191747
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr15_-_8191992 | 0.04 |
ENSDART00000155381
ENSDART00000190714 |
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr8_-_53044300 | 0.04 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr15_-_8177919 | 0.03 |
ENSDART00000101463
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr14_-_2221877 | 0.03 |
ENSDART00000106704
|
pcdh2ab1
|
protocadherin 2 alpha b 1 |
| chr7_-_13362590 | 0.03 |
ENSDART00000091616
|
sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr8_-_49201258 | 0.03 |
ENSDART00000114584
ENSDART00000142172 |
zgc:171501
|
zgc:171501 |
| chr4_-_53047883 | 0.03 |
ENSDART00000157496
|
si:dkey-201g16.5
|
si:dkey-201g16.5 |
| chr24_-_36723116 | 0.03 |
ENSDART00000088204
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr16_+_29586004 | 0.03 |
ENSDART00000149520
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr7_+_4971938 | 0.03 |
ENSDART00000172889
ENSDART00000145389 |
si:dkey-81n2.2
|
si:dkey-81n2.2 |
| chr17_-_5610514 | 0.03 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr8_-_8426504 | 0.03 |
ENSDART00000191686
ENSDART00000181523 |
cdk16
|
cyclin-dependent kinase 16 |
| chr12_-_44122412 | 0.02 |
ENSDART00000169094
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr10_+_40700311 | 0.02 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr1_-_11616142 | 0.02 |
ENSDART00000131668
|
si:dkey-26i13.5
|
si:dkey-26i13.5 |
| chr3_+_23710839 | 0.02 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr20_+_45853154 | 0.02 |
ENSDART00000181109
|
AL929237.4
|
|
| chr14_-_17305657 | 0.02 |
ENSDART00000168853
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr15_-_8191841 | 0.02 |
ENSDART00000156663
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr16_+_24972826 | 0.02 |
ENSDART00000063115
|
fpr1
|
formyl peptide receptor 1 |
| chr4_-_77377596 | 0.02 |
ENSDART00000186068
|
slco1e1
|
solute carrier organic anion transporter family, member 1E1 |
| chr19_+_3840955 | 0.01 |
ENSDART00000172305
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr9_-_3149896 | 0.01 |
ENSDART00000020861
|
pdk1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr13_-_29420885 | 0.01 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr16_-_29480335 | 0.01 |
ENSDART00000148930
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr18_-_20560007 | 0.01 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr5_-_56513825 | 0.01 |
ENSDART00000024207
|
tbx2a
|
T-box 2a |
| chr15_-_31406093 | 0.01 |
ENSDART00000123444
|
or111-8
|
odorant receptor, family D, subfamily 111, member 8 |
| chr4_-_46946643 | 0.01 |
ENSDART00000180392
ENSDART00000184563 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr1_+_8314826 | 0.01 |
ENSDART00000160440
|
cacna1hb
|
calcium channel, voltage-dependent, T type, alpha 1H subunit b |
| chr3_+_52684556 | 0.01 |
ENSDART00000139037
|
pgls
|
6-phosphogluconolactonase |
| chr17_-_50472864 | 0.01 |
ENSDART00000161066
ENSDART00000156963 |
si:ch211-235i11.3
|
si:ch211-235i11.3 |
| chr5_+_27897504 | 0.00 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr14_-_17305849 | 0.00 |
ENSDART00000160971
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxo3b+foxo6a+foxo6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 1.0 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.4 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 0.4 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.7 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.8 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.5 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.3 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 1.0 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.4 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 0.2 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.1 | 1.0 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.5 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.3 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.5 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.6 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 1.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.0 | GO:0005795 | Golgi stack(GO:0005795) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.7 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 1.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.4 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.1 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.5 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 1.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0032405 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 1.0 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |