Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for foxn1
Z-value: 0.43
Transcription factors associated with foxn1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxn1
|
ENSDARG00000011879 | forkhead box N1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxn1 | dr11_v1_chr15_-_28148314_28148314 | -0.37 | 1.4e-01 | Click! |
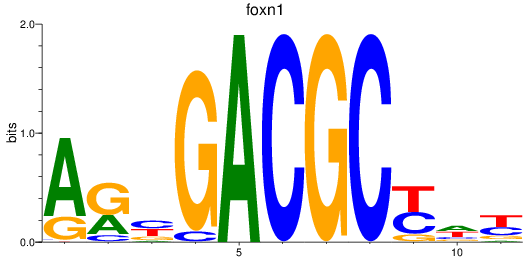
Activity profile of foxn1 motif
Sorted Z-values of foxn1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_29794058 | 1.11 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr3_+_7763114 | 1.11 |
ENSDART00000057434
|
hook2
|
hook microtubule-tethering protein 2 |
| chr8_+_16758304 | 0.96 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr15_-_37104165 | 0.92 |
ENSDART00000165867
|
zmp:0000001114
|
zmp:0000001114 |
| chr22_-_817479 | 0.92 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr22_-_38459316 | 0.88 |
ENSDART00000149683
ENSDART00000098461 |
ptk7a
|
protein tyrosine kinase 7a |
| chr11_-_23687158 | 0.88 |
ENSDART00000189599
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr14_+_22132896 | 0.84 |
ENSDART00000138274
|
ccng1
|
cyclin G1 |
| chr9_+_21793565 | 0.83 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr9_-_41090048 | 0.77 |
ENSDART00000131681
ENSDART00000182552 |
asnsd1
|
asparagine synthetase domain containing 1 |
| chr17_-_53022822 | 0.71 |
ENSDART00000103434
|
zgc:154061
|
zgc:154061 |
| chr20_-_13625588 | 0.70 |
ENSDART00000078893
|
sytl3
|
synaptotagmin-like 3 |
| chr8_-_25846188 | 0.69 |
ENSDART00000128829
|
efhd2
|
EF-hand domain family, member D2 |
| chr10_-_15854743 | 0.68 |
ENSDART00000092343
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr3_+_58472305 | 0.67 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr5_-_54554583 | 0.66 |
ENSDART00000158865
ENSDART00000158069 |
ssna1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr18_-_39787040 | 0.66 |
ENSDART00000169916
|
dmxl2
|
Dmx-like 2 |
| chr2_-_42492201 | 0.65 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr19_+_7173613 | 0.64 |
ENSDART00000001331
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr5_-_52216170 | 0.64 |
ENSDART00000158542
ENSDART00000192981 |
lnpep
|
leucyl/cystinyl aminopeptidase |
| chr20_-_6196989 | 0.63 |
ENSDART00000013343
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr12_-_10674606 | 0.62 |
ENSDART00000157919
|
med24
|
mediator complex subunit 24 |
| chr13_-_21650404 | 0.61 |
ENSDART00000078352
|
tspan14
|
tetraspanin 14 |
| chr21_-_4849029 | 0.61 |
ENSDART00000168930
ENSDART00000151019 |
notch1a
|
notch 1a |
| chr25_+_7532811 | 0.61 |
ENSDART00000161593
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr23_+_19590006 | 0.60 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr21_+_18405585 | 0.60 |
ENSDART00000139318
|
si:dkey-1d7.3
|
si:dkey-1d7.3 |
| chr14_-_46198373 | 0.60 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr3_-_18805225 | 0.59 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr4_+_23117557 | 0.58 |
ENSDART00000066909
|
slc35e3
|
solute carrier family 35, member E3 |
| chr2_-_2096055 | 0.58 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr23_+_6586467 | 0.58 |
ENSDART00000081763
ENSDART00000121480 |
rbm38
|
RNA binding motif protein 38 |
| chr21_-_35325466 | 0.58 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr19_-_6193448 | 0.56 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr15_+_34963316 | 0.56 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr2_+_42871831 | 0.56 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr21_+_21743599 | 0.56 |
ENSDART00000101700
|
pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr17_+_24064014 | 0.55 |
ENSDART00000182782
ENSDART00000139063 ENSDART00000132755 |
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr17_-_23895026 | 0.55 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr16_-_51072406 | 0.55 |
ENSDART00000083777
|
ago3a
|
argonaute RISC catalytic component 3a |
| chr23_-_10786400 | 0.54 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr25_-_34845302 | 0.52 |
ENSDART00000039485
|
gabarapl2
|
GABA(A) receptor-associated protein like 2 |
| chr9_+_54984900 | 0.51 |
ENSDART00000191622
|
mospd2
|
motile sperm domain containing 2 |
| chr14_+_22132388 | 0.51 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr10_+_29137482 | 0.49 |
ENSDART00000178280
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr20_+_51730658 | 0.49 |
ENSDART00000010271
|
aida
|
axin interactor, dorsalization associated |
| chr7_+_22801465 | 0.49 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr9_-_41090343 | 0.49 |
ENSDART00000187769
ENSDART00000180078 ENSDART00000166785 |
asnsd1
|
asparagine synthetase domain containing 1 |
| chr9_-_46415847 | 0.49 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr16_-_31351419 | 0.49 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr12_-_25150239 | 0.48 |
ENSDART00000038415
ENSDART00000135368 |
rhoq
|
ras homolog family member Q |
| chr7_+_24153070 | 0.47 |
ENSDART00000076735
|
lrp10
|
low density lipoprotein receptor-related protein 10 |
| chr6_-_7686594 | 0.47 |
ENSDART00000091836
ENSDART00000151697 |
ubn2a
|
ubinuclein 2a |
| chr9_+_54984537 | 0.47 |
ENSDART00000029528
|
mospd2
|
motile sperm domain containing 2 |
| chr5_+_23045096 | 0.46 |
ENSDART00000171719
|
atrxl
|
alpha thalassemia/mental retardation syndrome X-linked, like |
| chr19_-_6193067 | 0.46 |
ENSDART00000092656
ENSDART00000140347 |
erf
|
Ets2 repressor factor |
| chr7_+_24006875 | 0.45 |
ENSDART00000033755
|
homezb
|
homeobox and leucine zipper encoding b |
| chr6_+_40992409 | 0.45 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr2_+_16696052 | 0.45 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr16_+_23531583 | 0.44 |
ENSDART00000146708
|
adar
|
adenosine deaminase, RNA-specific |
| chr7_+_40083601 | 0.44 |
ENSDART00000099046
|
zgc:112356
|
zgc:112356 |
| chr7_-_60351876 | 0.44 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr16_+_50741154 | 0.44 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr6_+_59832786 | 0.43 |
ENSDART00000154985
ENSDART00000102148 |
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr21_+_21279159 | 0.43 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr15_-_34878388 | 0.43 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr10_+_3428194 | 0.43 |
ENSDART00000081599
|
ptpn11a
|
protein tyrosine phosphatase, non-receptor type 11, a |
| chr10_+_36695597 | 0.42 |
ENSDART00000169015
ENSDART00000171392 |
rab6a
|
RAB6A, member RAS oncogene family |
| chr7_-_60351537 | 0.42 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr6_+_12504462 | 0.42 |
ENSDART00000171460
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr5_-_19932621 | 0.42 |
ENSDART00000088881
|
git2a
|
G protein-coupled receptor kinase interacting ArfGAP 2a |
| chr10_+_375042 | 0.42 |
ENSDART00000171854
|
si:ch1073-303d10.1
|
si:ch1073-303d10.1 |
| chr6_-_42377307 | 0.41 |
ENSDART00000129302
|
emc3
|
ER membrane protein complex subunit 3 |
| chr25_+_7532627 | 0.41 |
ENSDART00000187660
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr17_-_16324565 | 0.40 |
ENSDART00000030835
|
hmbox1a
|
homeobox containing 1a |
| chr16_-_2390931 | 0.40 |
ENSDART00000149463
|
hace1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr13_+_32454262 | 0.39 |
ENSDART00000057421
|
rdh14a
|
retinol dehydrogenase 14a |
| chr10_+_29138021 | 0.39 |
ENSDART00000025227
ENSDART00000123033 ENSDART00000034242 |
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr17_+_44030692 | 0.39 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr13_+_2442841 | 0.38 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr14_+_22457230 | 0.38 |
ENSDART00000019296
|
gdf9
|
growth differentiation factor 9 |
| chr23_+_30898013 | 0.38 |
ENSDART00000146859
|
cables2a
|
Cdk5 and Abl enzyme substrate 2a |
| chr16_-_31505802 | 0.37 |
ENSDART00000147616
|
si:ch211-251p5.5
|
si:ch211-251p5.5 |
| chr18_+_39106722 | 0.37 |
ENSDART00000122377
|
bcl2l10
|
BCL2 like 10 |
| chr7_+_34236238 | 0.36 |
ENSDART00000052474
|
tipin
|
timeless interacting protein |
| chr11_+_18183220 | 0.36 |
ENSDART00000113468
|
LO018315.10
|
|
| chr25_-_25142387 | 0.36 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr8_+_23639124 | 0.34 |
ENSDART00000083108
|
nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr2_+_38055529 | 0.34 |
ENSDART00000145642
|
si:rp71-1g18.1
|
si:rp71-1g18.1 |
| chr5_+_63288599 | 0.34 |
ENSDART00000140065
|
si:ch73-37h15.2
|
si:ch73-37h15.2 |
| chr16_-_27161410 | 0.34 |
ENSDART00000177503
|
LO017771.1
|
|
| chr8_+_36503797 | 0.34 |
ENSDART00000184785
|
slc7a4
|
solute carrier family 7, member 4 |
| chr17_-_8656155 | 0.34 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr19_+_9232676 | 0.34 |
ENSDART00000136957
|
kmt2ba
|
lysine (K)-specific methyltransferase 2Ba |
| chr24_-_41267184 | 0.33 |
ENSDART00000063504
|
xylb
|
xylulokinase homolog (H. influenzae) |
| chr7_+_66884291 | 0.33 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr15_+_19335646 | 0.33 |
ENSDART00000062569
|
acad8
|
acyl-CoA dehydrogenase family, member 8 |
| chr4_-_12102025 | 0.33 |
ENSDART00000048391
ENSDART00000023894 |
braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr6_-_3998199 | 0.33 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr19_-_46088429 | 0.32 |
ENSDART00000161385
|
ptdss1b
|
phosphatidylserine synthase 1b |
| chr7_+_66884570 | 0.32 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr22_-_10156581 | 0.32 |
ENSDART00000168304
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr3_-_29891456 | 0.31 |
ENSDART00000151677
ENSDART00000014021 |
slc25a39
|
solute carrier family 25, member 39 |
| chr18_+_16715864 | 0.31 |
ENSDART00000079758
|
eif4g2b
|
eukaryotic translation initiation factor 4, gamma 2b |
| chr8_+_36500061 | 0.31 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr24_+_19210001 | 0.31 |
ENSDART00000179373
ENSDART00000139299 |
zgc:162928
|
zgc:162928 |
| chr10_-_76352 | 0.30 |
ENSDART00000186560
ENSDART00000144722 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr24_+_35387517 | 0.30 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr19_-_2861444 | 0.30 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr21_+_26028947 | 0.30 |
ENSDART00000028007
|
supt6h
|
SPT6 homolog, histone chaperone |
| chr16_-_48430272 | 0.30 |
ENSDART00000005927
|
rad21a
|
RAD21 cohesin complex component a |
| chr10_-_35177257 | 0.30 |
ENSDART00000077426
|
pom121
|
POM121 transmembrane nucleoporin |
| chr16_+_20871021 | 0.30 |
ENSDART00000006429
|
hibadhb
|
3-hydroxyisobutyrate dehydrogenase b |
| chr8_+_1284784 | 0.29 |
ENSDART00000061663
|
fbxl17
|
F-box and leucine-rich repeat protein 17 |
| chr25_+_3549841 | 0.29 |
ENSDART00000164030
|
ccdc77
|
coiled-coil domain containing 77 |
| chr13_-_25750910 | 0.29 |
ENSDART00000111567
|
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr5_-_52215926 | 0.29 |
ENSDART00000163973
ENSDART00000193602 |
lnpep
|
leucyl/cystinyl aminopeptidase |
| chr21_+_1119046 | 0.29 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr1_+_11107688 | 0.29 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr16_+_26547152 | 0.28 |
ENSDART00000141393
|
ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr15_-_47458034 | 0.28 |
ENSDART00000168527
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr5_-_30382925 | 0.28 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr6_+_34038963 | 0.28 |
ENSDART00000057732
ENSDART00000192496 |
ap1m3
|
adaptor-related protein complex 1, mu 3 subunit |
| chr19_-_29303788 | 0.28 |
ENSDART00000112167
|
srfbp1
|
serum response factor binding protein 1 |
| chr16_+_11242443 | 0.28 |
ENSDART00000024935
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr8_-_21071711 | 0.27 |
ENSDART00000111600
ENSDART00000135204 ENSDART00000131371 ENSDART00000146532 ENSDART00000137606 |
zgc:112962
|
zgc:112962 |
| chr8_-_21071476 | 0.27 |
ENSDART00000184184
ENSDART00000100288 |
zgc:112962
|
zgc:112962 |
| chr13_-_4018888 | 0.27 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr2_+_35732652 | 0.27 |
ENSDART00000052666
|
rasal2
|
RAS protein activator like 2 |
| chr21_-_3700334 | 0.27 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr9_-_52598343 | 0.26 |
ENSDART00000167922
|
xrcc5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 |
| chr16_+_48460873 | 0.26 |
ENSDART00000159902
|
ext1a
|
exostosin glycosyltransferase 1a |
| chr20_-_25445237 | 0.26 |
ENSDART00000145034
ENSDART00000018049 |
mrpl19
|
mitochondrial ribosomal protein L19 |
| chr1_+_44523516 | 0.26 |
ENSDART00000147702
|
zdhhc5a
|
zinc finger, DHHC-type containing 5a |
| chr6_-_45949121 | 0.26 |
ENSDART00000058555
|
tardbp
|
TAR DNA binding protein |
| chr8_+_52530889 | 0.25 |
ENSDART00000127729
ENSDART00000170360 ENSDART00000162687 |
stambpb
|
STAM binding protein b |
| chr7_+_13491452 | 0.25 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr25_+_7346800 | 0.25 |
ENSDART00000154404
|
peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr8_+_18010568 | 0.25 |
ENSDART00000121984
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr2_-_6292510 | 0.25 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr8_+_7778770 | 0.24 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr8_+_22277198 | 0.24 |
ENSDART00000005989
|
dffb
|
DNA fragmentation factor, beta polypeptide (caspase-activated DNase) |
| chr20_-_50014936 | 0.24 |
ENSDART00000148892
|
extl3
|
exostosin-like glycosyltransferase 3 |
| chr8_+_32722842 | 0.23 |
ENSDART00000147594
|
hmcn2
|
hemicentin 2 |
| chr16_-_13881139 | 0.23 |
ENSDART00000138904
ENSDART00000090221 ENSDART00000135099 |
cdc42ep5
|
CDC42 effector protein (Rho GTPase binding) 5 |
| chr1_-_25144439 | 0.23 |
ENSDART00000132355
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr7_+_69528850 | 0.23 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr25_+_2776511 | 0.23 |
ENSDART00000115280
|
neo1b
|
neogenin 1b |
| chr1_+_41690402 | 0.22 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr3_-_29891218 | 0.22 |
ENSDART00000142118
|
slc25a39
|
solute carrier family 25, member 39 |
| chr8_-_18010735 | 0.22 |
ENSDART00000125014
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr25_+_3549401 | 0.22 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr1_-_59313465 | 0.21 |
ENSDART00000158067
ENSDART00000159419 |
txndc11
|
thioredoxin domain containing 11 |
| chr25_-_16782394 | 0.21 |
ENSDART00000019413
|
ndufa9a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9a |
| chr8_+_18010978 | 0.21 |
ENSDART00000039887
ENSDART00000144532 |
ssbp3b
|
single stranded DNA binding protein 3b |
| chr16_-_6205790 | 0.21 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr3_+_15773991 | 0.20 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr19_-_7272921 | 0.20 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr12_-_4632519 | 0.20 |
ENSDART00000110514
|
prr12a
|
proline rich 12a |
| chr17_-_2584423 | 0.20 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr2_+_37480669 | 0.19 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr21_+_4256291 | 0.19 |
ENSDART00000148138
|
lrrc8aa
|
leucine rich repeat containing 8 VRAC subunit Aa |
| chr18_-_41160975 | 0.19 |
ENSDART00000187517
|
CABZ01005876.1
|
|
| chr13_-_51247529 | 0.19 |
ENSDART00000191774
ENSDART00000083788 |
AL929217.1
|
|
| chr4_-_4751981 | 0.19 |
ENSDART00000147436
ENSDART00000092984 ENSDART00000158466 |
creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr20_+_54333774 | 0.19 |
ENSDART00000144633
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr9_-_28399071 | 0.19 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr12_-_13318944 | 0.18 |
ENSDART00000152201
ENSDART00000041394 |
emc9
|
ER membrane protein complex subunit 9 |
| chr8_-_5220125 | 0.18 |
ENSDART00000035676
|
bnip3la
|
BCL2 interacting protein 3 like a |
| chr17_+_37301860 | 0.18 |
ENSDART00000181531
ENSDART00000075978 |
elmsan1b
|
ELM2 and Myb/SANT-like domain containing 1b |
| chr15_+_2559875 | 0.18 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr18_-_16937008 | 0.18 |
ENSDART00000100117
ENSDART00000022640 ENSDART00000136541 |
znf143b
|
zinc finger protein 143b |
| chr13_+_18321140 | 0.17 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr20_-_28884800 | 0.17 |
ENSDART00000134564
ENSDART00000132127 ENSDART00000135506 ENSDART00000075515 |
srsf5b
|
serine/arginine-rich splicing factor 5b |
| chr13_-_25284716 | 0.17 |
ENSDART00000039828
|
vcla
|
vinculin a |
| chr13_+_2448251 | 0.17 |
ENSDART00000188361
|
arfgef3
|
ARFGEF family member 3 |
| chr10_+_18877362 | 0.17 |
ENSDART00000138334
|
ppp2r2ab
|
protein phosphatase 2, regulatory subunit B, alpha b |
| chr12_+_3723650 | 0.17 |
ENSDART00000179922
ENSDART00000152482 ENSDART00000108771 |
pagr1
|
PAXIP1 associated glutamate-rich protein 1 |
| chr10_-_1180314 | 0.17 |
ENSDART00000114261
|
bmpr1bb
|
bone morphogenetic protein receptor, type IBb |
| chr3_+_56574623 | 0.17 |
ENSDART00000130877
|
rac1b
|
Rac family small GTPase 1b |
| chr17_-_32370047 | 0.17 |
ENSDART00000145487
|
klf11b
|
Kruppel-like factor 11b |
| chr21_-_42876565 | 0.17 |
ENSDART00000126480
|
zmp:0000001268
|
zmp:0000001268 |
| chr7_-_19068539 | 0.16 |
ENSDART00000108787
|
ndufs2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2 |
| chr5_+_51248784 | 0.16 |
ENSDART00000159571
ENSDART00000189513 ENSDART00000092285 |
msh3
|
mutS homolog 3 (E. coli) |
| chr2_-_32386791 | 0.16 |
ENSDART00000056634
|
ubtfl
|
upstream binding transcription factor, like |
| chr10_-_35108683 | 0.16 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr12_+_19320657 | 0.16 |
ENSDART00000100075
ENSDART00000066389 |
tmem184ba
|
transmembrane protein 184ba |
| chr10_+_37500234 | 0.16 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr5_-_35200590 | 0.15 |
ENSDART00000051271
|
fcho2
|
FCH domain only 2 |
| chr11_+_23933016 | 0.15 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr4_-_287425 | 0.15 |
ENSDART00000159128
|
echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr5_-_18962794 | 0.15 |
ENSDART00000145210
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr4_-_16883051 | 0.15 |
ENSDART00000124627
|
strap
|
serine/threonine kinase receptor associated protein |
| chr13_+_1439152 | 0.15 |
ENSDART00000159047
|
si:ch211-165e15.1
|
si:ch211-165e15.1 |
| chr5_+_38886499 | 0.15 |
ENSDART00000076845
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr19_-_10295173 | 0.14 |
ENSDART00000136697
|
cnot3b
|
CCR4-NOT transcription complex, subunit 3b |
| chr13_-_25284379 | 0.14 |
ENSDART00000124167
|
vcla
|
vinculin a |
| chr18_+_45645357 | 0.14 |
ENSDART00000010256
|
eif3m
|
eukaryotic translation initiation factor 3, subunit M |
| chr17_+_34186632 | 0.14 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr18_+_619619 | 0.14 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxn1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.2 | 0.7 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.2 | 0.8 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 0.6 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.2 | 0.6 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 1.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.4 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 0.4 | GO:0030237 | female sex determination(GO:0030237) |
| 0.1 | 0.3 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.1 | 0.9 | GO:2000095 | cerebrospinal fluid circulation(GO:0090660) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.5 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 0.5 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.1 | 0.5 | GO:0046329 | negative regulation of JUN kinase activity(GO:0043508) negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.1 | 1.2 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.1 | 0.2 | GO:1901004 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.1 | 0.2 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.3 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.1 | 0.4 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.5 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.1 | 0.3 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.3 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.1 | 0.2 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.4 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.0 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.3 | GO:0090024 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.6 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.2 | GO:0006921 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.2 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.6 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.2 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 1.1 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 1.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.0 | GO:2000257 | complement activation, alternative pathway(GO:0006957) regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.5 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.2 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.6 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.9 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.5 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.3 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 1.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0031464 | inclusion body(GO:0016234) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.8 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 0.6 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 0.6 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 1.0 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.3 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.3 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.5 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 1.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.0 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.2 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.0 | 0.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.8 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 1.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.6 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 1.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.2 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |