Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for foxm1
Z-value: 0.42
Transcription factors associated with foxm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxm1
|
ENSDARG00000003200 | forkhead box M1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxm1 | dr11_v1_chr4_-_5831522_5831522 | 0.75 | 3.5e-04 | Click! |
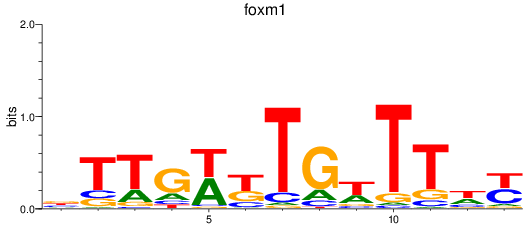
Activity profile of foxm1 motif
Sorted Z-values of foxm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_30039331 | 1.36 |
ENSDART00000086935
ENSDART00000143800 |
MARCH4 (1 of many)
|
zgc:153256 |
| chr14_-_52480661 | 0.87 |
ENSDART00000158353
|
exosc3
|
exosome component 3 |
| chr12_-_2993095 | 0.79 |
ENSDART00000152316
|
si:dkey-202c14.3
|
si:dkey-202c14.3 |
| chr7_+_24115082 | 0.70 |
ENSDART00000182718
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr16_+_2433696 | 0.63 |
ENSDART00000182803
|
CU928063.1
|
|
| chr19_+_31904836 | 0.63 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr5_-_13251907 | 0.60 |
ENSDART00000176774
ENSDART00000030553 |
top3b
|
DNA topoisomerase III beta |
| chr20_-_39596338 | 0.59 |
ENSDART00000023531
|
hey2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr19_+_43604643 | 0.58 |
ENSDART00000151168
|
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr2_-_32505091 | 0.58 |
ENSDART00000141884
ENSDART00000056639 |
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr7_+_24114694 | 0.57 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr19_+_11978209 | 0.57 |
ENSDART00000111568
|
polr2k
|
polymerase (RNA) II (DNA directed) polypeptide K |
| chr19_+_43604256 | 0.56 |
ENSDART00000151080
ENSDART00000110305 |
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr10_-_26512742 | 0.55 |
ENSDART00000135951
|
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr14_+_45565891 | 0.55 |
ENSDART00000133389
ENSDART00000025549 |
zgc:92249
|
zgc:92249 |
| chr7_-_41554047 | 0.54 |
ENSDART00000174144
|
plxdc2
|
plexin domain containing 2 |
| chr20_-_22798794 | 0.53 |
ENSDART00000148084
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr4_-_6809323 | 0.52 |
ENSDART00000099467
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr5_-_54672763 | 0.51 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr3_-_13461056 | 0.45 |
ENSDART00000137678
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr6_+_16949735 | 0.44 |
ENSDART00000155342
|
pimr13
|
Pim proto-oncogene, serine/threonine kinase, related 13 |
| chr11_+_2416064 | 0.44 |
ENSDART00000067117
|
ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr15_+_23784842 | 0.44 |
ENSDART00000192889
ENSDART00000138375 |
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr22_+_31059919 | 0.44 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr25_+_27873836 | 0.44 |
ENSDART00000163801
|
iqub
|
IQ motif and ubiquitin domain containing |
| chr5_-_68826177 | 0.43 |
ENSDART00000136605
|
si:ch211-283h6.4
|
si:ch211-283h6.4 |
| chr15_-_47822597 | 0.43 |
ENSDART00000193236
ENSDART00000161391 |
CZQB01095947.1
|
|
| chr8_+_40210398 | 0.43 |
ENSDART00000167612
|
rnf34a
|
ring finger protein 34a |
| chr7_+_52766211 | 0.42 |
ENSDART00000186191
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr16_-_24194587 | 0.42 |
ENSDART00000181520
|
rps19
|
ribosomal protein S19 |
| chr8_+_17987215 | 0.42 |
ENSDART00000113605
|
lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr15_-_23692359 | 0.41 |
ENSDART00000141618
|
ercc2
|
excision repair cross-complementation group 2 |
| chr3_-_16068236 | 0.41 |
ENSDART00000157315
|
si:dkey-81l17.6
|
si:dkey-81l17.6 |
| chr13_+_24679674 | 0.41 |
ENSDART00000033090
ENSDART00000139854 |
zgc:66426
|
zgc:66426 |
| chr17_+_46818521 | 0.41 |
ENSDART00000156022
|
pimr14
|
Pim proto-oncogene, serine/threonine kinase, related 14 |
| chr7_+_26629084 | 0.40 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr8_+_50531709 | 0.40 |
ENSDART00000193352
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr21_-_3796461 | 0.40 |
ENSDART00000021976
|
nsa2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr25_+_27873671 | 0.39 |
ENSDART00000088817
|
iqub
|
IQ motif and ubiquitin domain containing |
| chr6_-_16894312 | 0.39 |
ENSDART00000155450
|
pimr42
|
Pim proto-oncogene, serine/threonine kinase, related 42 |
| chr25_+_36045072 | 0.38 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr20_-_14925281 | 0.38 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr5_+_34982318 | 0.37 |
ENSDART00000160504
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr4_-_6567355 | 0.37 |
ENSDART00000134820
ENSDART00000142087 |
foxp2
|
forkhead box P2 |
| chr1_+_38362412 | 0.37 |
ENSDART00000075086
|
cep44
|
centrosomal protein 44 |
| chr4_-_75157223 | 0.36 |
ENSDART00000174127
|
CABZ01066312.1
|
|
| chr3_+_37112693 | 0.36 |
ENSDART00000055228
ENSDART00000144278 ENSDART00000138079 |
psmc3ip
|
PSMC3 interacting protein |
| chr14_-_33454595 | 0.36 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr24_+_5840258 | 0.36 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr7_-_51102479 | 0.36 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr4_-_17725008 | 0.35 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr20_+_30939178 | 0.35 |
ENSDART00000062556
|
sod2
|
superoxide dismutase 2, mitochondrial |
| chr14_-_4177311 | 0.34 |
ENSDART00000128129
|
si:dkey-185e18.7
|
si:dkey-185e18.7 |
| chr17_+_10593398 | 0.33 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr19_+_233143 | 0.33 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr2_+_22694382 | 0.32 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr10_-_35051691 | 0.31 |
ENSDART00000108670
ENSDART00000190711 |
supt20
|
SPT20 homolog, SAGA complex component |
| chr2_+_17540720 | 0.31 |
ENSDART00000114638
|
pimr198
|
Pim proto-oncogene, serine/threonine kinase, related 198 |
| chr6_-_16735402 | 0.31 |
ENSDART00000154216
|
pimr44
|
Pim proto-oncogene, serine/threonine kinase, related 44 |
| chr7_+_39401388 | 0.31 |
ENSDART00000144750
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr14_-_44790817 | 0.31 |
ENSDART00000098640
|
grhpra
|
glyoxylate reductase/hydroxypyruvate reductase a |
| chr21_+_13182149 | 0.31 |
ENSDART00000140267
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr17_+_46935061 | 0.31 |
ENSDART00000156578
|
pimr26
|
Pim proto-oncogene, serine/threonine kinase, related 26 |
| chr16_-_50203058 | 0.30 |
ENSDART00000154570
|
vsig10l
|
V-set and immunoglobulin domain containing 10 like |
| chr23_+_13928346 | 0.30 |
ENSDART00000155326
|
si:dkey-90a13.10
|
si:dkey-90a13.10 |
| chr10_+_35554219 | 0.30 |
ENSDART00000077373
|
zdhhc20a
|
zinc finger, DHHC-type containing 20a |
| chr15_+_37589698 | 0.29 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr13_+_12761707 | 0.29 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr20_+_16881883 | 0.29 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr10_-_35052198 | 0.29 |
ENSDART00000147805
|
supt20
|
SPT20 homolog, SAGA complex component |
| chr22_-_17606575 | 0.29 |
ENSDART00000183951
|
gpx4a
|
glutathione peroxidase 4a |
| chr12_-_48566778 | 0.28 |
ENSDART00000063442
|
cyp4f3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr5_-_64883082 | 0.28 |
ENSDART00000064983
ENSDART00000139066 |
krt1-c5
|
keratin, type 1, gene c5 |
| chr3_-_13461361 | 0.28 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr12_+_30234209 | 0.28 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr11_-_22369303 | 0.28 |
ENSDART00000163450
|
tmem183a
|
transmembrane protein 183A |
| chr11_+_6881001 | 0.28 |
ENSDART00000170331
|
klhl26
|
kelch-like family member 26 |
| chr5_+_34981584 | 0.27 |
ENSDART00000134795
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr2_-_10338759 | 0.27 |
ENSDART00000150166
ENSDART00000149584 |
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr10_+_19017146 | 0.26 |
ENSDART00000038674
|
tmem230a
|
transmembrane protein 230a |
| chr20_+_9124369 | 0.26 |
ENSDART00000064150
|
si:ch211-59d15.9
|
si:ch211-59d15.9 |
| chr3_-_37758487 | 0.26 |
ENSDART00000150938
|
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr9_-_38368138 | 0.26 |
ENSDART00000059574
|
ccdc93
|
coiled-coil domain containing 93 |
| chr3_+_5575313 | 0.26 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr22_+_2315996 | 0.26 |
ENSDART00000132489
|
znf1175
|
zinc finger protein 1175 |
| chr7_+_30282389 | 0.26 |
ENSDART00000108782
|
polr2m
|
RNA polymerase II subunit M |
| chr2_+_17524278 | 0.25 |
ENSDART00000165633
|
pimr196
|
Pim proto-oncogene, serine/threonine kinase, related 196 |
| chr22_-_9934854 | 0.25 |
ENSDART00000136404
|
si:dkey-253d23.11
|
si:dkey-253d23.11 |
| chr7_+_17255705 | 0.25 |
ENSDART00000053357
ENSDART00000167849 ENSDART00000165833 |
nitr9
si:ch73-46n24.1
|
novel immune-type receptor 9 si:ch73-46n24.1 |
| chr9_-_31108285 | 0.25 |
ENSDART00000003193
|
gpr183a
|
G protein-coupled receptor 183a |
| chr11_-_1509773 | 0.24 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr6_-_29288155 | 0.24 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr2_+_3595333 | 0.24 |
ENSDART00000041052
|
c1ql3b
|
complement component 1, q subcomponent-like 3b |
| chr17_+_43890736 | 0.24 |
ENSDART00000179918
|
msh4
|
mutS homolog 4 |
| chr4_-_890220 | 0.24 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr5_-_67757188 | 0.24 |
ENSDART00000167168
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr14_-_8453192 | 0.23 |
ENSDART00000136947
|
eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr21_-_25565392 | 0.23 |
ENSDART00000144917
ENSDART00000180102 |
si:dkey-17e16.10
|
si:dkey-17e16.10 |
| chr12_-_9516981 | 0.23 |
ENSDART00000106285
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| chr8_+_7033049 | 0.23 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr2_-_22688651 | 0.23 |
ENSDART00000013863
|
agxtb
|
alanine-glyoxylate aminotransferase b |
| chr14_+_23687678 | 0.22 |
ENSDART00000002469
|
hspa4b
|
heat shock protein 4b |
| chr11_-_28614608 | 0.22 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr17_-_46933567 | 0.22 |
ENSDART00000157274
|
pimr25
|
Pim proto-oncogene, serine/threonine kinase, related 25 |
| chr14_+_45559268 | 0.22 |
ENSDART00000173152
|
RARRES3
|
zgc:154040 |
| chr6_+_16870004 | 0.21 |
ENSDART00000154794
|
pimr21
|
Pim proto-oncogene, serine/threonine kinase, related 21 |
| chr6_+_18569453 | 0.21 |
ENSDART00000171338
|
rhot1b
|
ras homolog family member T1 |
| chr2_+_3986083 | 0.21 |
ENSDART00000188979
|
mkxb
|
mohawk homeobox b |
| chr4_-_22472653 | 0.21 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr18_-_15932704 | 0.21 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr11_-_24347644 | 0.21 |
ENSDART00000089777
|
si:ch211-15p9.2
|
si:ch211-15p9.2 |
| chr7_+_26716321 | 0.20 |
ENSDART00000189750
|
cd82a
|
CD82 molecule a |
| chr25_-_35045250 | 0.20 |
ENSDART00000156508
ENSDART00000126590 |
zgc:114046
|
zgc:114046 |
| chr5_-_24333684 | 0.20 |
ENSDART00000051553
|
znf703
|
zinc finger protein 703 |
| chr18_-_16795262 | 0.20 |
ENSDART00000048722
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr22_+_1421212 | 0.20 |
ENSDART00000161813
|
zgc:101130
|
zgc:101130 |
| chr4_+_25558849 | 0.19 |
ENSDART00000113663
ENSDART00000100755 ENSDART00000111416 ENSDART00000127840 ENSDART00000168618 ENSDART00000111820 ENSDART00000113866 ENSDART00000110107 ENSDART00000111344 ENSDART00000108548 |
zgc:195175
|
zgc:195175 |
| chr5_-_16351306 | 0.19 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr11_+_11201096 | 0.19 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr23_-_40817792 | 0.19 |
ENSDART00000136343
|
si:dkeyp-27c8.1
|
si:dkeyp-27c8.1 |
| chr23_+_39611688 | 0.19 |
ENSDART00000034690
|
otud3
|
OTU deubiquitinase 3 |
| chr13_+_13770980 | 0.18 |
ENSDART00000113089
|
slc4a11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr16_-_53800047 | 0.18 |
ENSDART00000158047
|
znrf2b
|
zinc and ring finger 2b |
| chr5_-_16475682 | 0.18 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr4_-_18436899 | 0.18 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr9_+_23770666 | 0.18 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr4_-_12388535 | 0.18 |
ENSDART00000017180
|
rergla
|
RERG/RAS-like a |
| chr18_+_19120984 | 0.18 |
ENSDART00000141501
|
si:dkey-242h9.3
|
si:dkey-242h9.3 |
| chr12_+_9542124 | 0.17 |
ENSDART00000127952
|
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr11_-_33618612 | 0.17 |
ENSDART00000033980
|
lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr19_+_7759354 | 0.17 |
ENSDART00000151400
|
ubap2l
|
ubiquitin associated protein 2-like |
| chr23_+_25305431 | 0.17 |
ENSDART00000143291
|
RPL41
|
si:dkey-151g10.6 |
| chr19_+_18627100 | 0.17 |
ENSDART00000167245
|
vps52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr10_+_35257651 | 0.17 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr18_-_37407235 | 0.17 |
ENSDART00000132315
|
cep126
|
centrosomal protein 126 |
| chr7_+_10592152 | 0.17 |
ENSDART00000182624
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr7_+_13995792 | 0.17 |
ENSDART00000091470
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr4_-_39111612 | 0.17 |
ENSDART00000150394
|
si:dkey-122c11.8
|
si:dkey-122c11.8 |
| chr2_-_29761965 | 0.17 |
ENSDART00000142057
ENSDART00000110595 |
si:dkey-188g12.1
|
si:dkey-188g12.1 |
| chr4_-_23759192 | 0.16 |
ENSDART00000014685
ENSDART00000131690 |
cpm
|
carboxypeptidase M |
| chr15_+_3674765 | 0.16 |
ENSDART00000081802
|
NEK3
|
NIMA related kinase 3 |
| chr6_+_17113677 | 0.16 |
ENSDART00000154080
|
pimr5
|
Pim proto-oncogene, serine/threonine kinase, related 5 |
| chr2_+_31476065 | 0.16 |
ENSDART00000049219
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr15_-_23784600 | 0.16 |
ENSDART00000059354
|
rad1
|
RAD1 homolog (S. pombe) |
| chr8_-_37056622 | 0.16 |
ENSDART00000111513
|
si:ch211-218o21.4
|
si:ch211-218o21.4 |
| chr16_+_53125918 | 0.16 |
ENSDART00000102170
|
CABZ01053976.1
|
|
| chr20_-_26383368 | 0.16 |
ENSDART00000024518
ENSDART00000087844 |
esr1
|
estrogen receptor 1 |
| chr5_+_26079178 | 0.16 |
ENSDART00000145920
|
si:dkey-201c13.2
|
si:dkey-201c13.2 |
| chr2_-_37837472 | 0.16 |
ENSDART00000165347
|
mettl17
|
methyltransferase like 17 |
| chr17_-_53440284 | 0.16 |
ENSDART00000126976
|
mycbp
|
c-myc binding protein |
| chr22_+_1734981 | 0.16 |
ENSDART00000158195
|
znf1159
|
zinc finger protein 1159 |
| chr14_+_52481288 | 0.15 |
ENSDART00000169164
ENSDART00000159297 |
tcerg1a
|
transcription elongation regulator 1a (CA150) |
| chr15_-_28082310 | 0.15 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr24_-_17444067 | 0.15 |
ENSDART00000155843
|
cntnap2a
|
contactin associated protein like 2a |
| chr6_+_17181157 | 0.15 |
ENSDART00000154962
|
pimr4
|
Pim proto-oncogene, serine/threonine kinase, related 4 |
| chr21_+_13353263 | 0.15 |
ENSDART00000114677
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr4_-_73787702 | 0.14 |
ENSDART00000136328
ENSDART00000150546 |
si:dkey-262g12.3
|
si:dkey-262g12.3 |
| chr6_-_23931442 | 0.14 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr11_+_6422374 | 0.14 |
ENSDART00000183148
|
FO681393.1
|
|
| chr13_-_280827 | 0.14 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr20_+_38525567 | 0.14 |
ENSDART00000147787
|
znf512
|
zinc finger protein 512 |
| chr6_-_16948040 | 0.13 |
ENSDART00000156433
|
pimr30
|
Pim proto-oncogene, serine/threonine kinase, related 30 |
| chr5_+_50898849 | 0.13 |
ENSDART00000083317
|
arsk
|
arylsulfatase family, member K |
| chr7_+_38811800 | 0.13 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr9_+_2522797 | 0.13 |
ENSDART00000186786
ENSDART00000147034 |
gpr155a
|
G protein-coupled receptor 155a |
| chr17_-_53439866 | 0.13 |
ENSDART00000154826
|
mycbp
|
c-myc binding protein |
| chr14_+_31865099 | 0.13 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr14_+_8343498 | 0.13 |
ENSDART00000164551
|
nrg2b
|
neuregulin 2b |
| chr11_+_44502410 | 0.12 |
ENSDART00000172998
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr16_-_55028740 | 0.12 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr6_+_16736871 | 0.12 |
ENSDART00000155471
|
pimr12
|
Pim proto-oncogene, serine/threonine kinase, related 12 |
| chr6_-_16804001 | 0.12 |
ENSDART00000155398
|
pimr40
|
Pim proto-oncogene, serine/threonine kinase, related 40 |
| chr14_+_22498757 | 0.12 |
ENSDART00000021657
|
smyd5
|
SMYD family member 5 |
| chr8_+_32719930 | 0.12 |
ENSDART00000145362
|
hmcn2
|
hemicentin 2 |
| chr14_-_26177156 | 0.12 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr11_-_42418374 | 0.12 |
ENSDART00000160704
|
slmapa
|
sarcolemma associated protein a |
| chr6_+_35362225 | 0.11 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr1_-_23268013 | 0.11 |
ENSDART00000146575
|
rfc1
|
replication factor C (activator 1) 1 |
| chr19_-_18626952 | 0.11 |
ENSDART00000168004
ENSDART00000162034 ENSDART00000165486 ENSDART00000167971 |
rps18
|
ribosomal protein S18 |
| chr7_-_50604626 | 0.11 |
ENSDART00000073903
ENSDART00000174031 |
crtc3
|
CREB regulated transcription coactivator 3 |
| chr5_-_38197080 | 0.11 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr24_-_2312868 | 0.11 |
ENSDART00000140125
ENSDART00000138432 |
cul2
|
cullin 2 |
| chr21_+_3796620 | 0.11 |
ENSDART00000099535
ENSDART00000144515 |
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr4_-_68913650 | 0.10 |
ENSDART00000184297
|
si:dkey-264f17.5
|
si:dkey-264f17.5 |
| chr7_-_20836625 | 0.10 |
ENSDART00000192566
|
cldn15a
|
claudin 15a |
| chr15_-_23761580 | 0.10 |
ENSDART00000137918
|
bbc3
|
BCL2 binding component 3 |
| chr9_-_38036984 | 0.10 |
ENSDART00000134574
|
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr22_+_2254972 | 0.10 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr6_+_17267681 | 0.10 |
ENSDART00000156312
|
pimr15
|
Pim proto-oncogene, serine/threonine kinase, related 15 |
| chr25_+_28679672 | 0.10 |
ENSDART00000139965
ENSDART00000134072 |
cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr20_-_39273987 | 0.09 |
ENSDART00000127173
|
clu
|
clusterin |
| chr25_+_4812685 | 0.09 |
ENSDART00000193103
|
myo5c
|
myosin VC |
| chr19_-_27261102 | 0.09 |
ENSDART00000143919
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr1_+_55535827 | 0.09 |
ENSDART00000152784
|
adgre16
|
adhesion G protein-coupled receptor E16 |
| chr22_-_29205327 | 0.09 |
ENSDART00000183161
ENSDART00000189515 |
pvalb7
|
parvalbumin 7 |
| chr11_-_25213651 | 0.09 |
ENSDART00000097316
ENSDART00000152186 |
myh7ba
|
myosin, heavy chain 7B, cardiac muscle, beta a |
| chr8_+_8973425 | 0.09 |
ENSDART00000066107
|
bcap31
|
B cell receptor associated protein 31 |
| chr7_+_1505507 | 0.09 |
ENSDART00000161015
|
nop10
|
NOP10 ribonucleoprotein homolog (yeast) |
| chr7_+_60359347 | 0.09 |
ENSDART00000145201
ENSDART00000039827 |
ppp1r14bb
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Bb |
| chr12_+_19191787 | 0.09 |
ENSDART00000152892
|
slc16a8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0071034 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.2 | 0.6 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.1 | 0.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.4 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 0.3 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.3 | GO:0032757 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.2 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 1.1 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 0.4 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.4 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.1 | 0.2 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.0 | 0.4 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.3 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.4 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0071450 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 1.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.4 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.3 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.1 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0090497 | skin morphogenesis(GO:0043589) mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.3 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.1 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 3.6 | GO:0046777 | protein autophosphorylation(GO:0046777) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.9 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.4 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 0.4 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 1.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 1.3 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.8 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.4 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.0 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 2.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |