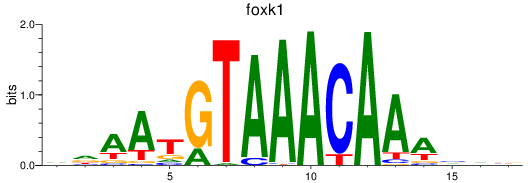
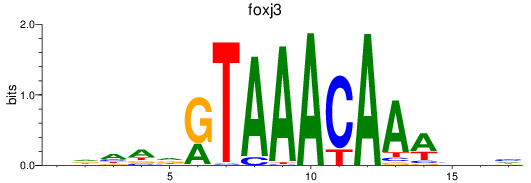
Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for foxk1_foxj3
Z-value: 1.03
Transcription factors associated with foxk1_foxj3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxk1
|
ENSDARG00000037872 | forkhead box K1 |
|
foxj3
|
ENSDARG00000075774 | forkhead box J3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxk1 | dr11_v1_chr3_-_40933415_40933415 | 0.59 | 1.0e-02 | Click! |
| foxj3 | dr11_v1_chr11_-_26362294_26362334 | 0.26 | 2.9e-01 | Click! |
Activity profile of foxk1_foxj3 motif
Sorted Z-values of foxk1_foxj3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_21295132 | 4.12 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr9_+_52411530 | 3.30 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr5_+_44944778 | 3.26 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr25_+_13662606 | 3.18 |
ENSDART00000008989
|
ccdc113
|
coiled-coil domain containing 113 |
| chr5_-_54672763 | 2.80 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr16_-_49646625 | 2.30 |
ENSDART00000101629
|
efhb
|
EF-hand domain family, member B |
| chr1_+_50538839 | 2.05 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr10_+_39084354 | 1.95 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr14_+_26439227 | 1.94 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr25_-_35360096 | 1.86 |
ENSDART00000154053
ENSDART00000171917 |
si:ch73-147o17.1
|
si:ch73-147o17.1 |
| chr12_+_19188542 | 1.79 |
ENSDART00000134726
ENSDART00000148011 ENSDART00000109541 |
cby1
|
chibby homolog 1 (Drosophila) |
| chr24_-_6678640 | 1.75 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr14_-_30960470 | 1.61 |
ENSDART00000129989
|
si:ch211-191o15.6
|
si:ch211-191o15.6 |
| chr16_-_6944927 | 1.59 |
ENSDART00000149620
|
pmvk
|
phosphomevalonate kinase |
| chr1_-_44901163 | 1.57 |
ENSDART00000145354
|
tcirg1a
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3a |
| chr7_+_20110336 | 1.47 |
ENSDART00000179395
|
zgc:114045
|
zgc:114045 |
| chr4_-_12795030 | 1.45 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr20_-_13640598 | 1.42 |
ENSDART00000128823
ENSDART00000103394 |
rsph3
|
radial spoke 3 homolog |
| chr25_+_19149241 | 1.38 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr18_+_26899316 | 1.37 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr11_+_25257022 | 1.34 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr2_-_11662851 | 1.34 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr5_-_42661012 | 1.24 |
ENSDART00000158339
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr5_+_49744713 | 1.22 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr4_-_12795436 | 1.22 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr5_+_42400777 | 1.20 |
ENSDART00000183114
|
BX548073.8
|
|
| chr16_+_6944564 | 1.16 |
ENSDART00000104252
|
eaf1
|
ELL associated factor 1 |
| chr13_+_35635672 | 1.15 |
ENSDART00000148481
|
thbs2a
|
thrombospondin 2a |
| chr24_+_35787629 | 1.11 |
ENSDART00000136721
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr17_+_24632440 | 1.07 |
ENSDART00000157092
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr2_+_49572059 | 1.06 |
ENSDART00000108861
|
sema4e
|
semaphorin 4e |
| chr17_+_4325693 | 1.03 |
ENSDART00000154264
|
mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr24_+_17269849 | 1.03 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr5_+_26913120 | 0.98 |
ENSDART00000126609
|
tbx3b
|
T-box 3b |
| chr14_-_17576391 | 0.97 |
ENSDART00000161355
ENSDART00000168959 |
rnf4
|
ring finger protein 4 |
| chr16_-_41131578 | 0.96 |
ENSDART00000102649
ENSDART00000145956 |
ptpn23a
|
protein tyrosine phosphatase, non-receptor type 23, a |
| chr4_-_6373735 | 0.94 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr8_-_23783633 | 0.92 |
ENSDART00000132657
|
si:ch211-163l21.7
|
si:ch211-163l21.7 |
| chr21_-_13661631 | 0.90 |
ENSDART00000184408
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr8_+_37755099 | 0.90 |
ENSDART00000075708
ENSDART00000140966 |
got1l1
|
glutamic-oxaloacetic transaminase 1 like 1 |
| chr25_-_3470910 | 0.89 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr21_+_5800306 | 0.88 |
ENSDART00000020603
|
ccng2
|
cyclin G2 |
| chr12_-_11457625 | 0.87 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr14_-_36799280 | 0.87 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr21_+_28502340 | 0.85 |
ENSDART00000077897
ENSDART00000140229 |
otub1a
|
OTU deubiquitinase, ubiquitin aldehyde binding 1a |
| chr4_-_22472653 | 0.83 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr24_-_23758003 | 0.81 |
ENSDART00000178085
|
BX640462.2
|
Danio rerio minichromosome maintenance domain containing 2 (mcmdc2), mRNA. |
| chr23_-_21957383 | 0.81 |
ENSDART00000145780
|
si:dkey-68l7.2
|
si:dkey-68l7.2 |
| chr21_-_32301109 | 0.81 |
ENSDART00000139890
|
clk4b
|
CDC-like kinase 4b |
| chr3_+_24618012 | 0.80 |
ENSDART00000111997
|
zgc:171506
|
zgc:171506 |
| chr12_+_34763795 | 0.80 |
ENSDART00000153097
|
slc38a10
|
solute carrier family 38, member 10 |
| chr16_+_6944311 | 0.79 |
ENSDART00000144763
|
eaf1
|
ELL associated factor 1 |
| chr4_-_17725008 | 0.79 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr15_+_25681044 | 0.79 |
ENSDART00000077853
|
hic1
|
hypermethylated in cancer 1 |
| chr18_+_21122818 | 0.79 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr2_+_19633493 | 0.79 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr10_+_6641514 | 0.78 |
ENSDART00000150003
|
si:ch211-57m13.5
|
si:ch211-57m13.5 |
| chr19_-_32600823 | 0.78 |
ENSDART00000134149
ENSDART00000187858 |
zgc:91944
|
zgc:91944 |
| chr2_+_55365727 | 0.77 |
ENSDART00000162943
|
FP245456.1
|
|
| chr1_-_9940494 | 0.77 |
ENSDART00000138726
|
tmem8a
|
transmembrane protein 8A |
| chr3_-_60711127 | 0.77 |
ENSDART00000184119
|
ubald2
|
UBA-like domain containing 2 |
| chr5_-_40190949 | 0.76 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr12_+_8822717 | 0.74 |
ENSDART00000021628
|
reep3b
|
receptor accessory protein 3b |
| chr3_+_19207176 | 0.74 |
ENSDART00000087803
|
rln3a
|
relaxin 3a |
| chr13_+_22119798 | 0.73 |
ENSDART00000173206
ENSDART00000078652 ENSDART00000165842 |
camk2g2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 2 |
| chr2_+_24786765 | 0.73 |
ENSDART00000141030
|
pde4ca
|
phosphodiesterase 4C, cAMP-specific a |
| chr16_+_28578648 | 0.71 |
ENSDART00000149566
|
nmt2
|
N-myristoyltransferase 2 |
| chr21_-_28920245 | 0.70 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr11_+_6819050 | 0.69 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr23_+_24085531 | 0.68 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr19_-_28367413 | 0.68 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr21_-_11855828 | 0.68 |
ENSDART00000081666
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr5_-_4931266 | 0.67 |
ENSDART00000067600
|
zbtb43
|
zinc finger and BTB domain containing 43 |
| chr14_-_25599002 | 0.67 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr24_-_31306724 | 0.66 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr8_+_4803906 | 0.65 |
ENSDART00000045533
|
tmem127
|
transmembrane protein 127 |
| chr11_+_6295370 | 0.65 |
ENSDART00000139882
|
ranbp3a
|
RAN binding protein 3a |
| chr9_-_33785093 | 0.65 |
ENSDART00000140779
ENSDART00000059837 |
fundc1
|
FUN14 domain containing 1 |
| chr5_-_38384289 | 0.64 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr14_+_94946 | 0.64 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr7_-_13906409 | 0.64 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr21_-_11856143 | 0.63 |
ENSDART00000151204
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr6_-_39160422 | 0.63 |
ENSDART00000148661
|
stat2
|
signal transducer and activator of transcription 2 |
| chr16_-_12097558 | 0.62 |
ENSDART00000123142
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr22_+_4707663 | 0.62 |
ENSDART00000042194
|
cers4a
|
ceramide synthase 4a |
| chr13_+_22675802 | 0.61 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr13_+_9368621 | 0.60 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr11_+_16153207 | 0.59 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr7_-_24828296 | 0.59 |
ENSDART00000138193
|
otub1b
|
OTU deubiquitinase, ubiquitin aldehyde binding 1b |
| chr11_+_16152316 | 0.58 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr10_+_2529037 | 0.58 |
ENSDART00000123467
|
zmp:0000001301
|
zmp:0000001301 |
| chr13_+_28854438 | 0.57 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr7_+_52761841 | 0.57 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr8_-_30791266 | 0.56 |
ENSDART00000062220
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr3_-_20063671 | 0.56 |
ENSDART00000056614
ENSDART00000126915 |
ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr25_+_18953756 | 0.55 |
ENSDART00000154291
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr23_-_28025943 | 0.54 |
ENSDART00000181146
|
sp5l
|
Sp5 transcription factor-like |
| chr14_+_36226243 | 0.54 |
ENSDART00000052569
|
pitx2
|
paired-like homeodomain 2 |
| chr16_-_12097394 | 0.53 |
ENSDART00000103944
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr3_-_45298487 | 0.53 |
ENSDART00000102245
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr13_-_23665580 | 0.53 |
ENSDART00000144282
|
map3k21
|
mitogen-activated protein kinase kinase kinase 21 |
| chr9_+_2343096 | 0.52 |
ENSDART00000062292
ENSDART00000191722 ENSDART00000135180 |
atf2
|
activating transcription factor 2 |
| chr2_-_19576640 | 0.52 |
ENSDART00000141021
|
pimr51
|
Pim proto-oncogene, serine/threonine kinase, related 51 |
| chr17_-_13026634 | 0.52 |
ENSDART00000113713
|
fam177a1
|
family with sequence similarity 177, member A1 |
| chr17_+_43843536 | 0.52 |
ENSDART00000164097
|
FO704777.1
|
|
| chr9_-_7640692 | 0.52 |
ENSDART00000135616
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr25_+_18953575 | 0.51 |
ENSDART00000155610
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr15_+_3284684 | 0.50 |
ENSDART00000179778
|
foxo1a
|
forkhead box O1 a |
| chr4_-_14926637 | 0.50 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr20_-_19590378 | 0.49 |
ENSDART00000152588
|
baalcb
|
brain and acute leukemia, cytoplasmic b |
| chr14_-_38843690 | 0.49 |
ENSDART00000183629
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr16_+_29492937 | 0.49 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr25_+_3677650 | 0.49 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr8_+_8893535 | 0.49 |
ENSDART00000139150
|
otud5a
|
OTU deubiquitinase 5a |
| chr25_-_11088839 | 0.48 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr15_+_3284416 | 0.47 |
ENSDART00000187665
ENSDART00000171723 |
foxo1a
|
forkhead box O1 a |
| chr18_-_48550426 | 0.47 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr4_-_20135919 | 0.47 |
ENSDART00000172230
|
cep83
|
centrosomal protein 83 |
| chr8_-_30791089 | 0.47 |
ENSDART00000147332
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr17_-_2690083 | 0.47 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr5_+_13549603 | 0.47 |
ENSDART00000171270
|
ckap2l
|
cytoskeleton associated protein 2-like |
| chr6_-_43792179 | 0.46 |
ENSDART00000160849
|
foxp1b
|
forkhead box P1b |
| chr20_+_52774730 | 0.45 |
ENSDART00000014606
|
phactr1
|
phosphatase and actin regulator 1 |
| chr16_-_24605969 | 0.45 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr6_-_9565526 | 0.45 |
ENSDART00000151470
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr17_+_32500387 | 0.44 |
ENSDART00000018423
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr16_+_29492749 | 0.44 |
ENSDART00000179680
|
ctsk
|
cathepsin K |
| chr21_+_30502002 | 0.43 |
ENSDART00000043727
|
slc7a3a
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3a |
| chr23_-_30787932 | 0.43 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr19_-_19339285 | 0.42 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr7_+_22849657 | 0.42 |
ENSDART00000173483
|
pygmb
|
phosphorylase, glycogen, muscle b |
| chr25_+_18954189 | 0.42 |
ENSDART00000123207
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr20_-_23171430 | 0.42 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr11_+_18612421 | 0.42 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr9_+_38163876 | 0.42 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr11_-_12051805 | 0.41 |
ENSDART00000110117
ENSDART00000182744 |
socs7
|
suppressor of cytokine signaling 7 |
| chr21_+_11415224 | 0.40 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr4_-_20135406 | 0.40 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr19_-_4851411 | 0.40 |
ENSDART00000110398
|
fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr4_+_7391110 | 0.40 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr3_+_20012891 | 0.39 |
ENSDART00000078974
|
tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr1_+_25801648 | 0.39 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr9_+_14291932 | 0.39 |
ENSDART00000183119
|
BX511082.1
|
|
| chr12_+_30234209 | 0.38 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr8_-_29822527 | 0.37 |
ENSDART00000167487
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr7_-_51102479 | 0.36 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr6_-_30839763 | 0.36 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr1_-_30689004 | 0.36 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr20_+_25552057 | 0.36 |
ENSDART00000102913
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr14_-_33177935 | 0.35 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr15_-_36533322 | 0.35 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr5_-_23485161 | 0.34 |
ENSDART00000170293
ENSDART00000134069 |
si:dkeyp-20g2.1
si:dkeyp-20g2.3
|
si:dkeyp-20g2.1 si:dkeyp-20g2.3 |
| chr23_-_16682186 | 0.33 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr21_-_280769 | 0.33 |
ENSDART00000157753
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr5_+_36611128 | 0.32 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr11_-_21586157 | 0.32 |
ENSDART00000190095
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr25_+_17920361 | 0.32 |
ENSDART00000185644
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr1_+_41886830 | 0.32 |
ENSDART00000185887
ENSDART00000084735 |
si:dkey-37g12.1
|
si:dkey-37g12.1 |
| chr21_-_20765338 | 0.32 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr3_+_55031685 | 0.31 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr16_-_19373310 | 0.31 |
ENSDART00000148294
|
dnah11
|
dynein, axonemal, heavy chain 11 |
| chr14_-_34044369 | 0.31 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr15_+_23784842 | 0.31 |
ENSDART00000192889
ENSDART00000138375 |
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr19_-_32600638 | 0.31 |
ENSDART00000143497
|
zgc:91944
|
zgc:91944 |
| chr21_-_13662237 | 0.30 |
ENSDART00000091647
ENSDART00000151547 |
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr20_-_1314537 | 0.30 |
ENSDART00000144288
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr16_-_15263099 | 0.30 |
ENSDART00000125691
|
sntb1
|
syntrophin, basic 1 |
| chr20_+_40457599 | 0.30 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr21_-_22951604 | 0.30 |
ENSDART00000083449
ENSDART00000180129 |
dub
|
duboraya |
| chr11_+_7264457 | 0.29 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr17_+_8324345 | 0.29 |
ENSDART00000172443
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr13_-_45155284 | 0.29 |
ENSDART00000161144
|
runx3
|
runt-related transcription factor 3 |
| chr10_+_31951338 | 0.29 |
ENSDART00000019416
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr15_+_19990068 | 0.29 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr3_-_16039619 | 0.28 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr1_+_53945934 | 0.28 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr8_+_23093155 | 0.28 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr2_-_8017579 | 0.28 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr23_-_27608257 | 0.27 |
ENSDART00000026314
|
phf8
|
PHD finger protein 8 |
| chr13_+_18471546 | 0.27 |
ENSDART00000090712
ENSDART00000127962 |
stox1
|
storkhead box 1 |
| chr2_+_38161318 | 0.27 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr2_-_54054225 | 0.27 |
ENSDART00000167239
|
CABZ01050249.1
|
|
| chr20_-_16849306 | 0.27 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr19_+_40337457 | 0.27 |
ENSDART00000089547
ENSDART00000140857 |
fam133b
|
family with sequence similarity 133, member B |
| chr13_+_45980163 | 0.26 |
ENSDART00000074547
ENSDART00000005195 |
bsdc1
|
BSD domain containing 1 |
| chr13_+_42602406 | 0.26 |
ENSDART00000133388
ENSDART00000147996 |
mlh1
|
mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) |
| chr25_-_16826219 | 0.26 |
ENSDART00000191299
ENSDART00000188504 |
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr4_-_49312747 | 0.26 |
ENSDART00000181057
|
BX942819.2
|
|
| chr16_+_43152727 | 0.25 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr12_-_32013125 | 0.25 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr9_+_41459759 | 0.25 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr1_+_52130213 | 0.24 |
ENSDART00000018817
|
rnf11a
|
ring finger protein 11a |
| chr12_-_22509069 | 0.24 |
ENSDART00000179755
ENSDART00000109707 |
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr24_+_11381400 | 0.24 |
ENSDART00000058703
|
ackr4b
|
atypical chemokine receptor 4b |
| chr19_-_10395683 | 0.23 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr5_-_16351306 | 0.23 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr6_-_8244474 | 0.22 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr22_+_26798853 | 0.22 |
ENSDART00000087576
ENSDART00000179780 |
slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr6_+_49021703 | 0.22 |
ENSDART00000149394
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr19_+_1878500 | 0.21 |
ENSDART00000113392
|
ggcta
|
gamma-glutamylcyclotransferase a |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxk1_foxj3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.7 | 2.1 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.6 | 1.9 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.5 | 4.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.4 | 1.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 1.4 | GO:1900044 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 0.7 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 0.9 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.2 | 1.1 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 0.8 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.2 | 2.7 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.2 | 0.7 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.1 | 0.4 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.1 | 1.0 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 1.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.1 | 0.4 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.6 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 1.2 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 1.4 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 1.6 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.5 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.3 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.4 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.5 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.1 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.3 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.6 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 2.0 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.1 | 0.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.3 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.1 | 0.3 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 0.9 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.7 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 0.4 | GO:1902837 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.3 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.4 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.6 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.5 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.2 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.4 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.0 | 1.0 | GO:0032392 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.1 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.0 | 1.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 1.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.3 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 1.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.3 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.5 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 1.1 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 1.0 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 1.1 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.3 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.5 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.1 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.8 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.9 | GO:0003205 | cardiac chamber development(GO:0003205) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.1 | GO:0044273 | sulfur compound catabolic process(GO:0044273) |
| 0.0 | 0.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 1.3 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.7 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.0 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.3 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.4 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.8 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 2.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 3.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 2.0 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 1.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 1.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 2.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.5 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 1.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.4 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 1.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 2.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 1.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 0.9 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 0.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 0.8 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.2 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.7 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.2 | 1.5 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.2 | 0.8 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 1.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.6 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 2.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.6 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 1.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 2.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.4 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.3 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.4 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 1.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.6 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 1.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 2.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.9 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |