Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
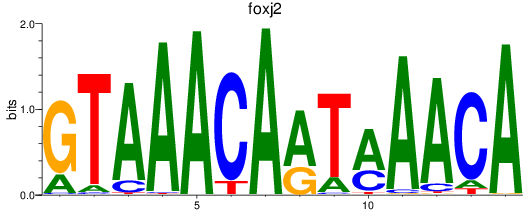
Results for foxj2
Z-value: 0.74
Transcription factors associated with foxj2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxj2
|
ENSDARG00000057680 | forkhead box J2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxj2 | dr11_v1_chr16_-_12787029_12787029 | -0.93 | 2.7e-08 | Click! |
Activity profile of foxj2 motif
Sorted Z-values of foxj2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_30285613 | 1.63 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr17_+_38602790 | 1.59 |
ENSDART00000062010
|
ccdc88c
|
coiled-coil domain containing 88C |
| chr16_+_54210554 | 1.50 |
ENSDART00000172622
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr14_+_21699129 | 1.47 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr10_+_15025006 | 1.46 |
ENSDART00000145192
ENSDART00000140084 |
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr23_-_27822920 | 1.37 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr15_-_33172246 | 1.36 |
ENSDART00000158666
|
nbeab
|
neurobeachin b |
| chr15_+_30126971 | 1.35 |
ENSDART00000100214
|
nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr10_-_35257458 | 1.34 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr22_-_718615 | 1.33 |
ENSDART00000149320
|
arl8a
|
ADP-ribosylation factor-like 8A |
| chr25_-_13789955 | 1.30 |
ENSDART00000167742
ENSDART00000165116 ENSDART00000171461 |
ckap5
|
cytoskeleton associated protein 5 |
| chr13_-_31687925 | 1.24 |
ENSDART00000085989
|
trmt5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae) |
| chr10_+_45031398 | 1.23 |
ENSDART00000160536
|
gnsb
|
glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID), b |
| chr18_+_7553950 | 1.22 |
ENSDART00000193420
ENSDART00000062143 |
zgc:77650
|
zgc:77650 |
| chr17_+_51743908 | 1.19 |
ENSDART00000149039
ENSDART00000148869 |
odc1
|
ornithine decarboxylase 1 |
| chr14_-_34633960 | 1.16 |
ENSDART00000128869
ENSDART00000179977 |
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr16_-_41646164 | 1.15 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr14_+_21699414 | 1.14 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr13_+_31687973 | 1.14 |
ENSDART00000076479
|
slc38a6
|
solute carrier family 38, member 6 |
| chr19_-_11846958 | 1.11 |
ENSDART00000148516
|
ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr1_-_14503182 | 1.10 |
ENSDART00000190599
|
si:dkey-194g4.1
|
si:dkey-194g4.1 |
| chr15_-_45356635 | 1.08 |
ENSDART00000192000
|
CABZ01068246.1
|
|
| chr16_-_41714988 | 1.08 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr20_+_25904199 | 1.06 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr19_-_15229421 | 1.00 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr11_+_37768298 | 1.00 |
ENSDART00000166886
|
sox13
|
SRY (sex determining region Y)-box 13 |
| chr19_-_34873566 | 0.99 |
ENSDART00000016057
|
ctnnal1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr4_+_13586455 | 0.97 |
ENSDART00000187230
|
tnpo3
|
transportin 3 |
| chr6_-_40449399 | 0.95 |
ENSDART00000103879
|
tatdn2
|
TatD DNase domain containing 2 |
| chr16_+_33143503 | 0.93 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr15_-_4415917 | 0.93 |
ENSDART00000062874
|
atp1b3b
|
ATPase Na+/K+ transporting subunit beta 3b |
| chr10_+_37173029 | 0.92 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr5_-_9678075 | 0.89 |
ENSDART00000097217
|
FAM109B
|
si:ch211-193c2.2 |
| chr20_+_33924235 | 0.88 |
ENSDART00000146292
ENSDART00000139609 |
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr19_-_25119443 | 0.86 |
ENSDART00000148953
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr13_-_377256 | 0.85 |
ENSDART00000137398
|
ch1073-291c23.1
|
ch1073-291c23.1 |
| chr17_+_15534815 | 0.84 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr17_+_33415319 | 0.83 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr16_+_39146696 | 0.83 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr2_-_42492201 | 0.82 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr5_+_63302660 | 0.82 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr19_-_35492693 | 0.81 |
ENSDART00000135838
ENSDART00000051745 ENSDART00000177052 |
ptp4a2b
|
protein tyrosine phosphatase type IVA, member 2b |
| chr17_+_33415542 | 0.79 |
ENSDART00000183169
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr8_+_40284973 | 0.79 |
ENSDART00000018401
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr3_+_34159192 | 0.78 |
ENSDART00000151119
|
carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr3_+_13862753 | 0.77 |
ENSDART00000168315
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr19_+_46222918 | 0.77 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr19_+_6990970 | 0.77 |
ENSDART00000158758
ENSDART00000160482 ENSDART00000193566 |
kifc1
|
kinesin family member C1 |
| chr17_+_11372531 | 0.76 |
ENSDART00000130975
ENSDART00000149366 |
timm9
|
translocase of inner mitochondrial membrane 9 homolog |
| chr14_-_47963115 | 0.75 |
ENSDART00000003826
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr25_-_19608382 | 0.75 |
ENSDART00000022279
ENSDART00000135201 ENSDART00000147223 ENSDART00000190220 ENSDART00000184242 ENSDART00000166824 |
gtse1
|
G-2 and S-phase expressed 1 |
| chr10_-_42898220 | 0.74 |
ENSDART00000099270
|
CU326366.2
|
|
| chr19_+_23982466 | 0.74 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr2_+_22602301 | 0.74 |
ENSDART00000038514
|
sept2
|
septin 2 |
| chr13_+_39315881 | 0.74 |
ENSDART00000135999
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr19_+_46222428 | 0.73 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr23_-_17450746 | 0.72 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr18_+_17534627 | 0.71 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr3_-_55328548 | 0.68 |
ENSDART00000082944
|
dock6
|
dedicator of cytokinesis 6 |
| chr11_-_11878099 | 0.67 |
ENSDART00000188314
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr16_+_35916371 | 0.67 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr9_-_34842414 | 0.66 |
ENSDART00000126348
|
BX601644.1
|
Danio rerio cytokine receptor-like factor 2 (crlf2), mRNA. |
| chr4_+_13586689 | 0.66 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr22_+_4442473 | 0.66 |
ENSDART00000170751
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr10_+_11261576 | 0.66 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr17_+_23753975 | 0.65 |
ENSDART00000135561
|
zgc:91976
|
zgc:91976 |
| chr5_-_69212184 | 0.65 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr16_-_6198543 | 0.65 |
ENSDART00000167505
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr8_-_11202378 | 0.64 |
ENSDART00000147817
ENSDART00000174039 |
fam208b
|
family with sequence similarity 208, member B |
| chr16_+_43077909 | 0.63 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr7_+_67325933 | 0.62 |
ENSDART00000170575
ENSDART00000183342 |
nfat5b
|
nuclear factor of activated T cells 5b |
| chr15_-_5901514 | 0.62 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr21_-_34261677 | 0.61 |
ENSDART00000124649
ENSDART00000172381 ENSDART00000064320 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr3_-_31893008 | 0.59 |
ENSDART00000127297
|
ddx42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr16_+_13818500 | 0.59 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr13_-_869808 | 0.59 |
ENSDART00000189029
|
AL929536.6
|
|
| chr8_+_48942470 | 0.58 |
ENSDART00000005464
ENSDART00000132035 |
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr7_-_32782430 | 0.56 |
ENSDART00000173808
|
gas2b
|
growth arrest-specific 2b |
| chr1_+_17527931 | 0.56 |
ENSDART00000131927
|
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr16_-_20870143 | 0.55 |
ENSDART00000169541
ENSDART00000040727 |
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr1_-_36770883 | 0.55 |
ENSDART00000167831
|
prmt9
|
protein arginine methyltransferase 9 |
| chr4_+_9612574 | 0.54 |
ENSDART00000150336
ENSDART00000041289 ENSDART00000150828 |
tmem243b
|
transmembrane protein 243, mitochondrial b |
| chr4_+_14727018 | 0.54 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr15_-_18574716 | 0.53 |
ENSDART00000142010
ENSDART00000019006 |
ncam1b
|
neural cell adhesion molecule 1b |
| chr2_-_29996036 | 0.53 |
ENSDART00000020792
|
cnpy1
|
canopy1 |
| chr11_-_45420212 | 0.51 |
ENSDART00000182042
ENSDART00000163185 |
ankrd13c
|
ankyrin repeat domain 13C |
| chr25_-_6391271 | 0.50 |
ENSDART00000138782
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr3_+_59864872 | 0.49 |
ENSDART00000102014
|
mcrip1
|
MAPK regulated corepressor interacting protein 1 |
| chr13_+_30421472 | 0.49 |
ENSDART00000143569
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr4_+_14727212 | 0.49 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr17_+_24597001 | 0.48 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr7_+_69528850 | 0.46 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr23_-_41762956 | 0.46 |
ENSDART00000128302
|
stk35
|
serine/threonine kinase 35 |
| chr8_-_43750062 | 0.45 |
ENSDART00000142243
|
ulk1a
|
unc-51 like autophagy activating kinase 1a |
| chr18_+_20034023 | 0.45 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr6_+_12527725 | 0.44 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr19_+_4856351 | 0.44 |
ENSDART00000093402
|
cdk12
|
cyclin-dependent kinase 12 |
| chr23_-_27607039 | 0.44 |
ENSDART00000183639
|
phf8
|
PHD finger protein 8 |
| chr13_-_25476920 | 0.43 |
ENSDART00000131403
|
tial1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr8_+_23147609 | 0.43 |
ENSDART00000180284
|
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr15_-_7598294 | 0.42 |
ENSDART00000165898
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr8_+_17838924 | 0.41 |
ENSDART00000170762
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr15_-_7598542 | 0.41 |
ENSDART00000173092
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr5_-_63302944 | 0.40 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr16_+_45922175 | 0.39 |
ENSDART00000018253
|
rbm8a
|
RNA binding motif protein 8A |
| chr8_-_51954562 | 0.38 |
ENSDART00000132527
ENSDART00000057315 |
cep78
|
centrosomal protein 78 |
| chr16_+_44768361 | 0.38 |
ENSDART00000036302
|
upk1a
|
uroplakin 1a |
| chr24_+_8904741 | 0.37 |
ENSDART00000140924
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr19_-_45650994 | 0.36 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr1_-_49947290 | 0.35 |
ENSDART00000141476
|
sgms2
|
sphingomyelin synthase 2 |
| chr25_+_28563465 | 0.34 |
ENSDART00000186861
|
SLC15A5
|
si:ch211-190o6.3 |
| chr3_+_17878124 | 0.33 |
ENSDART00000166430
ENSDART00000163421 ENSDART00000121473 |
dnajc7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr5_+_53009083 | 0.32 |
ENSDART00000157836
|
hint2
|
histidine triad nucleotide binding protein 2 |
| chr23_-_41762797 | 0.32 |
ENSDART00000186564
|
stk35
|
serine/threonine kinase 35 |
| chr23_+_20640875 | 0.32 |
ENSDART00000147382
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr6_-_40063359 | 0.32 |
ENSDART00000157119
ENSDART00000059636 ENSDART00000156385 |
chchd4b
|
coiled-coil-helix-coiled-coil-helix domain containing 4b |
| chr18_+_41560822 | 0.32 |
ENSDART00000158503
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr10_-_1180314 | 0.30 |
ENSDART00000114261
|
bmpr1bb
|
bone morphogenetic protein receptor, type IBb |
| chr18_+_41561285 | 0.29 |
ENSDART00000169621
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr19_+_41551335 | 0.28 |
ENSDART00000169193
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr4_+_76671012 | 0.28 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr12_-_33706726 | 0.28 |
ENSDART00000153135
|
myo15b
|
myosin XVB |
| chr2_+_30480907 | 0.27 |
ENSDART00000041378
ENSDART00000138863 |
fam173b
|
family with sequence similarity 173, member B |
| chr13_-_10431476 | 0.26 |
ENSDART00000133968
|
camkmt
|
calmodulin-lysine N-methyltransferase |
| chr16_-_32975951 | 0.26 |
ENSDART00000101969
ENSDART00000175149 |
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr20_-_3390406 | 0.25 |
ENSDART00000136987
|
rev3l
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr16_-_18960333 | 0.25 |
ENSDART00000143185
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr15_-_26570948 | 0.25 |
ENSDART00000156621
|
wdr81
|
WD repeat domain 81 |
| chr3_-_29891456 | 0.25 |
ENSDART00000151677
ENSDART00000014021 |
slc25a39
|
solute carrier family 25, member 39 |
| chr22_-_5099824 | 0.24 |
ENSDART00000122341
ENSDART00000161345 |
zfr2
|
zinc finger RNA binding protein 2 |
| chr3_+_23045214 | 0.24 |
ENSDART00000157090
ENSDART00000156472 |
b4galnt2.2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 2 |
| chr19_+_18739085 | 0.24 |
ENSDART00000188868
|
spaca4l
|
sperm acrosome associated 4 like |
| chr6_-_14004772 | 0.24 |
ENSDART00000185629
|
zgc:92027
|
zgc:92027 |
| chr8_+_7097929 | 0.22 |
ENSDART00000188955
ENSDART00000184772 ENSDART00000109581 |
abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr5_-_37875636 | 0.22 |
ENSDART00000184674
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr13_+_22865676 | 0.21 |
ENSDART00000057651
|
hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr5_-_68259747 | 0.20 |
ENSDART00000188756
|
CABZ01083944.1
|
|
| chr8_-_10949847 | 0.20 |
ENSDART00000123209
|
pqlc2
|
PQ loop repeat containing 2 |
| chr9_-_32158288 | 0.20 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr3_-_29891218 | 0.20 |
ENSDART00000142118
|
slc25a39
|
solute carrier family 25, member 39 |
| chr1_-_55166511 | 0.20 |
ENSDART00000150430
ENSDART00000035725 |
pane1
|
proliferation associated nuclear element |
| chr25_+_3217419 | 0.19 |
ENSDART00000104859
|
rccd1
|
RCC1 domain containing 1 |
| chr1_-_45177373 | 0.19 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr2_+_29996650 | 0.19 |
ENSDART00000138050
ENSDART00000141026 |
rbm33b
|
RNA binding motif protein 33b |
| chr5_+_42957503 | 0.18 |
ENSDART00000192885
|
mob1ba
|
MOB kinase activator 1Ba |
| chr19_+_41551543 | 0.18 |
ENSDART00000112364
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr1_-_9940494 | 0.16 |
ENSDART00000138726
|
tmem8a
|
transmembrane protein 8A |
| chr18_-_14743659 | 0.16 |
ENSDART00000125979
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| chr4_-_19883985 | 0.16 |
ENSDART00000014440
|
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr8_+_7097740 | 0.16 |
ENSDART00000159670
|
abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr18_-_25855263 | 0.15 |
ENSDART00000042074
|
sema4ba
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ba |
| chr8_+_23521974 | 0.15 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr7_+_37372479 | 0.14 |
ENSDART00000173652
|
sall1a
|
spalt-like transcription factor 1a |
| chr16_+_10318893 | 0.14 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr7_-_59047720 | 0.13 |
ENSDART00000184831
|
CR376779.1
|
|
| chr2_-_42827336 | 0.12 |
ENSDART00000140913
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr17_+_16755287 | 0.12 |
ENSDART00000080129
|
ston2
|
stonin 2 |
| chr4_-_19884440 | 0.12 |
ENSDART00000182979
ENSDART00000105967 |
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr16_-_17560822 | 0.11 |
ENSDART00000089230
|
casp2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr15_+_21254800 | 0.11 |
ENSDART00000142902
|
usf1
|
upstream transcription factor 1 |
| chr2_+_3516913 | 0.11 |
ENSDART00000109346
|
CU693445.1
|
|
| chr2_-_24603325 | 0.10 |
ENSDART00000113356
|
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr15_+_42573909 | 0.09 |
ENSDART00000181801
|
CLDN8 (1 of many)
|
zgc:110333 |
| chr8_-_46457233 | 0.09 |
ENSDART00000113214
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr23_+_32335871 | 0.08 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr23_-_7594723 | 0.08 |
ENSDART00000115298
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr20_-_33966148 | 0.08 |
ENSDART00000148111
|
selp
|
selectin P |
| chr4_-_16353733 | 0.08 |
ENSDART00000186785
|
lum
|
lumican |
| chr19_-_2318391 | 0.08 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| chr12_+_4149112 | 0.07 |
ENSDART00000166191
|
itgam
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr21_+_45757317 | 0.07 |
ENSDART00000163152
|
h2afy
|
H2A histone family, member Y |
| chr1_-_29761086 | 0.07 |
ENSDART00000136760
|
alg11
|
asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| chr7_-_20464468 | 0.07 |
ENSDART00000134700
|
cnpy4
|
canopy4 |
| chr1_-_26444075 | 0.07 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr17_-_28797395 | 0.07 |
ENSDART00000134735
|
scfd1
|
sec1 family domain containing 1 |
| chr12_-_9790485 | 0.07 |
ENSDART00000027321
|
prdm9
|
PR domain containing 9 |
| chr9_+_12887491 | 0.06 |
ENSDART00000102386
|
si:ch211-167j6.4
|
si:ch211-167j6.4 |
| chr11_-_16394971 | 0.06 |
ENSDART00000180981
ENSDART00000179925 |
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr24_-_24881996 | 0.05 |
ENSDART00000145712
|
crhb
|
corticotropin releasing hormone b |
| chr2_+_15203322 | 0.05 |
ENSDART00000144171
|
abca4b
|
ATP-binding cassette, sub-family A (ABC1), member 4b |
| chr4_-_1908179 | 0.05 |
ENSDART00000139586
|
ano6
|
anoctamin 6 |
| chr5_+_11812089 | 0.04 |
ENSDART00000111359
|
fbxo21
|
F-box protein 21 |
| chr4_+_40858450 | 0.04 |
ENSDART00000152047
|
si:dkey-285e18.6
|
si:dkey-285e18.6 |
| chr10_+_34426571 | 0.04 |
ENSDART00000144529
|
nbeaa
|
neurobeachin a |
| chr19_-_3303995 | 0.03 |
ENSDART00000105150
|
si:ch211-133n4.9
|
si:ch211-133n4.9 |
| chr21_+_40225915 | 0.03 |
ENSDART00000048475
ENSDART00000174122 |
or115-12
|
odorant receptor, family F, subfamily 115, member 12 |
| chr14_+_16151368 | 0.03 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr11_+_28107632 | 0.03 |
ENSDART00000177721
|
nmur3
|
neuromedin U receptor 3 |
| chr17_+_5985933 | 0.02 |
ENSDART00000190844
|
zgc:194275
|
zgc:194275 |
| chr2_-_38337122 | 0.02 |
ENSDART00000076523
ENSDART00000187473 |
slc7a8b
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8b |
| chr15_-_29598444 | 0.02 |
ENSDART00000154847
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr9_+_19248124 | 0.02 |
ENSDART00000185312
|
htr1fa
|
5-hydroxytryptamine (serotonin) receptor 1Fa |
| chr4_-_75681004 | 0.02 |
ENSDART00000186296
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr9_-_32730487 | 0.02 |
ENSDART00000147795
|
olig1
|
oligodendrocyte transcription factor 1 |
| chr13_-_2010191 | 0.01 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr16_+_5259886 | 0.01 |
ENSDART00000186668
|
plecb
|
plectin b |
| chr18_+_6039141 | 0.01 |
ENSDART00000138972
|
si:ch73-386h18.1
|
si:ch73-386h18.1 |
| chr1_+_33322174 | 0.01 |
ENSDART00000140435
|
mxra5a
|
matrix-remodelling associated 5a |
| chr25_+_14870043 | 0.01 |
ENSDART00000035714
ENSDART00000171835 |
dnajc24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr14_+_16151636 | 0.01 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxj2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.4 | 1.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.3 | 1.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 1.2 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.3 | 1.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.2 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.2 | 0.7 | GO:0035666 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.2 | 0.7 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.2 | 0.8 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 0.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.2 | 0.6 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.2 | 1.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 1.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.4 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 2.4 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.1 | 1.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 1.4 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 1.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 1.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 1.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.6 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.3 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 1.6 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.7 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 0.3 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.9 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.8 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.6 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.5 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 1.5 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.8 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.4 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.7 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 2.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 1.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 1.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.2 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.4 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 1.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.8 | GO:0032432 | actin filament bundle(GO:0032432) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.2 | 1.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 0.8 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.2 | 1.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 0.7 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 0.8 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.1 | 1.2 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 1.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.4 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.7 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.0 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.6 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.6 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.3 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 4.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 0.4 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 1.0 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.9 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.7 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.2 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 3.9 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |