Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
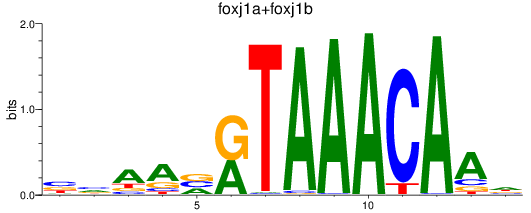
Results for foxj1a+foxj1b
Z-value: 1.09
Transcription factors associated with foxj1a+foxj1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxj1b
|
ENSDARG00000088290 | forkhead box J1b |
|
foxj1a
|
ENSDARG00000101919 | forkhead box J1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxj1b | dr11_v1_chr12_+_20149707_20149707 | 0.67 | 2.5e-03 | Click! |
| foxj1a | dr11_v1_chr3_+_60716904_60716904 | 0.66 | 3.0e-03 | Click! |
Activity profile of foxj1a+foxj1b motif
Sorted Z-values of foxj1a+foxj1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_49646625 | 3.39 |
ENSDART00000101629
|
efhb
|
EF-hand domain family, member B |
| chr12_+_17100021 | 3.14 |
ENSDART00000177923
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr5_+_39563301 | 3.04 |
ENSDART00000131245
ENSDART00000097872 |
si:ch211-57h10.1
|
si:ch211-57h10.1 |
| chr5_-_42661012 | 2.60 |
ENSDART00000158339
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr5_-_54672763 | 2.54 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr5_+_42400777 | 2.48 |
ENSDART00000183114
|
BX548073.8
|
|
| chr9_+_52411530 | 2.30 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr7_+_38395197 | 2.30 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr3_+_55031685 | 2.29 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr20_-_13640598 | 2.28 |
ENSDART00000128823
ENSDART00000103394 |
rsph3
|
radial spoke 3 homolog |
| chr19_+_5315987 | 2.19 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr15_+_25681044 | 2.07 |
ENSDART00000077853
|
hic1
|
hypermethylated in cancer 1 |
| chr25_-_10571078 | 2.00 |
ENSDART00000153898
|
si:ch211-107e6.5
|
si:ch211-107e6.5 |
| chr21_-_11855828 | 1.91 |
ENSDART00000081666
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr2_+_55365727 | 1.90 |
ENSDART00000162943
|
FP245456.1
|
|
| chr19_+_619200 | 1.89 |
ENSDART00000050125
|
nupl2
|
nucleoporin like 2 |
| chr7_-_33023404 | 1.86 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr21_-_11856143 | 1.83 |
ENSDART00000151204
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr23_+_17220986 | 1.82 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr23_+_33296588 | 1.79 |
ENSDART00000030368
|
si:ch211-226m16.3
|
si:ch211-226m16.3 |
| chr21_+_28502340 | 1.78 |
ENSDART00000077897
ENSDART00000140229 |
otub1a
|
OTU deubiquitinase, ubiquitin aldehyde binding 1a |
| chr1_+_53945934 | 1.75 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr18_+_21122818 | 1.74 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr1_+_55239160 | 1.71 |
ENSDART00000152318
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr12_-_10705916 | 1.70 |
ENSDART00000164038
|
FO704786.1
|
|
| chr6_+_43015916 | 1.68 |
ENSDART00000064888
|
tcta
|
T cell leukemia translocation altered |
| chr6_+_29791164 | 1.66 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr4_+_27100531 | 1.62 |
ENSDART00000115200
|
alg12
|
ALG12, alpha-1,6-mannosyltransferase |
| chr17_+_24632440 | 1.61 |
ENSDART00000157092
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr4_-_12795030 | 1.60 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr3_+_32671146 | 1.60 |
ENSDART00000039466
|
rnf25
|
ring finger protein 25 |
| chr4_-_17725008 | 1.59 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr20_-_1314537 | 1.57 |
ENSDART00000144288
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr25_-_37435060 | 1.53 |
ENSDART00000102855
ENSDART00000148566 |
pdhx
|
pyruvate dehydrogenase complex, component X |
| chr24_-_37640705 | 1.50 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr7_+_48805534 | 1.50 |
ENSDART00000145375
ENSDART00000148744 |
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr14_-_36799280 | 1.49 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr23_+_24085531 | 1.47 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr25_-_35360096 | 1.43 |
ENSDART00000154053
ENSDART00000171917 |
si:ch73-147o17.1
|
si:ch73-147o17.1 |
| chr2_+_19633493 | 1.43 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr20_-_2667902 | 1.42 |
ENSDART00000036373
|
cfap206
|
cilia and flagella associated protein 206 |
| chr10_-_3416258 | 1.40 |
ENSDART00000005168
|
ddx55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr4_-_64703 | 1.40 |
ENSDART00000167851
|
CU856344.1
|
|
| chr11_+_16153207 | 1.39 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr15_-_44465679 | 1.37 |
ENSDART00000054605
|
aasdhppt
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr19_-_32600823 | 1.36 |
ENSDART00000134149
ENSDART00000187858 |
zgc:91944
|
zgc:91944 |
| chr23_-_7755373 | 1.33 |
ENSDART00000162868
|
PCMTD2 (1 of many)
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr11_+_16152316 | 1.30 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr9_+_8898024 | 1.30 |
ENSDART00000134954
ENSDART00000143671 ENSDART00000147098 ENSDART00000179969 ENSDART00000111214 ENSDART00000140232 ENSDART00000139687 ENSDART00000132443 ENSDART00000130208 |
naxd
|
NAD(P)HX dehydratase |
| chr1_-_44901163 | 1.29 |
ENSDART00000145354
|
tcirg1a
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3a |
| chr20_-_1314355 | 1.27 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr3_-_45298487 | 1.25 |
ENSDART00000102245
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr15_+_23784842 | 1.22 |
ENSDART00000192889
ENSDART00000138375 |
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr4_-_12795436 | 1.22 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr21_-_5881344 | 1.21 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr15_+_6661343 | 1.20 |
ENSDART00000160136
|
nop53
|
NOP53 ribosome biogenesis factor |
| chr19_+_1878500 | 1.19 |
ENSDART00000113392
|
ggcta
|
gamma-glutamylcyclotransferase a |
| chr5_+_40299568 | 1.19 |
ENSDART00000142157
|
arl15a
|
ADP-ribosylation factor-like 15a |
| chr7_+_52761841 | 1.18 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr15_+_46386261 | 1.16 |
ENSDART00000191793
|
igsf11
|
immunoglobulin superfamily member 11 |
| chr13_-_25842074 | 1.16 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr14_-_17576391 | 1.16 |
ENSDART00000161355
ENSDART00000168959 |
rnf4
|
ring finger protein 4 |
| chr21_+_10577527 | 1.15 |
ENSDART00000165070
ENSDART00000127963 |
ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr20_-_48061351 | 1.14 |
ENSDART00000164962
|
prep
|
prolyl endopeptidase |
| chr11_-_16152400 | 1.13 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr16_-_24195252 | 1.13 |
ENSDART00000136205
ENSDART00000048599 ENSDART00000161547 |
rps19
|
ribosomal protein S19 |
| chr16_-_28856112 | 1.13 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr23_-_31266586 | 1.13 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr23_+_24598910 | 1.13 |
ENSDART00000126510
ENSDART00000078796 |
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr21_-_3796461 | 1.07 |
ENSDART00000021976
|
nsa2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr17_+_6667441 | 1.05 |
ENSDART00000123503
ENSDART00000180381 ENSDART00000156140 |
smc6
|
structural maintenance of chromosomes 6 |
| chr16_+_40576679 | 1.05 |
ENSDART00000169412
ENSDART00000193464 |
ccne2
|
cyclin E2 |
| chr16_-_50203058 | 1.05 |
ENSDART00000154570
|
vsig10l
|
V-set and immunoglobulin domain containing 10 like |
| chr20_+_15552657 | 1.05 |
ENSDART00000063912
|
jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr25_-_13188214 | 1.05 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr7_-_28549361 | 1.05 |
ENSDART00000173918
ENSDART00000054368 ENSDART00000113313 |
st5
|
suppression of tumorigenicity 5 |
| chr16_-_26731928 | 1.04 |
ENSDART00000135860
|
rnf41l
|
ring finger protein 41, like |
| chr21_+_11415224 | 1.04 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr2_+_19522082 | 1.03 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr2_+_14992879 | 1.03 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr5_-_20446082 | 1.03 |
ENSDART00000051607
|
ISCU (1 of many)
|
si:ch211-191d15.2 |
| chr17_+_53284040 | 1.02 |
ENSDART00000165368
|
atxn3
|
ataxin 3 |
| chr21_-_22951604 | 1.02 |
ENSDART00000083449
ENSDART00000180129 |
dub
|
duboraya |
| chr14_-_28566238 | 1.02 |
ENSDART00000172547
|
insb
|
preproinsulin b |
| chr9_-_2573121 | 1.01 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr14_-_28001986 | 1.01 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr14_-_21219659 | 1.00 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr9_+_31222026 | 1.00 |
ENSDART00000145573
|
clybl
|
citrate lyase beta like |
| chr20_+_26881600 | 1.00 |
ENSDART00000174799
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr3_-_40664868 | 0.99 |
ENSDART00000138783
ENSDART00000178567 |
rnf216
|
ring finger protein 216 |
| chr22_+_37631234 | 0.98 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr17_+_15297398 | 0.98 |
ENSDART00000156574
|
si:ch211-270g19.5
|
si:ch211-270g19.5 |
| chr11_-_6048490 | 0.97 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr2_-_19576640 | 0.97 |
ENSDART00000141021
|
pimr51
|
Pim proto-oncogene, serine/threonine kinase, related 51 |
| chr11_+_8660158 | 0.96 |
ENSDART00000169141
|
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr2_-_19520690 | 0.96 |
ENSDART00000133559
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr5_+_40485503 | 0.95 |
ENSDART00000051055
|
ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) |
| chr5_-_66160415 | 0.92 |
ENSDART00000073895
|
mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr16_-_13595027 | 0.90 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr5_+_44944778 | 0.90 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr19_-_5103313 | 0.90 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr2_+_19578079 | 0.89 |
ENSDART00000144413
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr14_+_30910114 | 0.88 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr11_+_7264457 | 0.88 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr24_-_31306724 | 0.88 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr10_+_35257651 | 0.87 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr12_+_18899396 | 0.87 |
ENSDART00000105858
|
xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr7_-_38807819 | 0.85 |
ENSDART00000132772
ENSDART00000052324 |
atg13
|
ATG13 autophagy related 13 homolog (S. cerevisiae) |
| chr19_-_5103141 | 0.83 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr15_+_36941490 | 0.83 |
ENSDART00000172664
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr20_+_16173618 | 0.83 |
ENSDART00000192109
ENSDART00000104112 ENSDART00000129633 |
zyg11
|
zyg-11 homolog (C. elegans) |
| chr2_-_19630297 | 0.83 |
ENSDART00000100111
ENSDART00000142173 |
pimr51
BX088713.2
|
Pim proto-oncogene, serine/threonine kinase, related 51 |
| chr19_-_19339285 | 0.83 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr24_+_37640626 | 0.83 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr15_-_17960228 | 0.83 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr2_-_31376606 | 0.83 |
ENSDART00000098988
ENSDART00000125746 |
clul1
|
clusterin-like 1 (retinal) |
| chr8_+_10305400 | 0.82 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr10_-_34089779 | 0.81 |
ENSDART00000140070
|
pimr144
|
Pim proto-oncogene, serine/threonine kinase, related 144 |
| chr12_-_32013125 | 0.81 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr23_-_30787932 | 0.80 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr18_-_48550426 | 0.80 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr9_-_2572790 | 0.80 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr9_-_19161982 | 0.80 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr25_-_31423493 | 0.79 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr16_-_41131578 | 0.78 |
ENSDART00000102649
ENSDART00000145956 |
ptpn23a
|
protein tyrosine phosphatase, non-receptor type 23, a |
| chr22_-_2959005 | 0.78 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr12_-_11457625 | 0.78 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr22_+_19553390 | 0.78 |
ENSDART00000061739
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr7_+_48805725 | 0.78 |
ENSDART00000166543
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr6_+_32834760 | 0.77 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr1_+_33668236 | 0.77 |
ENSDART00000122316
ENSDART00000102184 |
arl13b
|
ADP-ribosylation factor-like 13b |
| chr3_+_57038033 | 0.76 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr5_-_23317477 | 0.76 |
ENSDART00000090171
|
nlgn3b
|
neuroligin 3b |
| chr23_+_28077953 | 0.74 |
ENSDART00000186122
ENSDART00000111570 |
slc26a10
|
solute carrier family 26, member 10 |
| chr9_+_38163876 | 0.70 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr12_+_30234209 | 0.69 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr18_+_5875268 | 0.69 |
ENSDART00000177784
ENSDART00000122009 |
wdr59
|
WD repeat domain 59 |
| chr19_+_1606185 | 0.69 |
ENSDART00000092183
|
lrrc3b
|
leucine rich repeat containing 3B |
| chr20_+_6142433 | 0.68 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr7_-_37555208 | 0.67 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr24_+_11381400 | 0.67 |
ENSDART00000058703
|
ackr4b
|
atypical chemokine receptor 4b |
| chr24_+_25032340 | 0.67 |
ENSDART00000005845
|
mtmr6
|
myotubularin related protein 6 |
| chr21_-_28920245 | 0.67 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr12_-_8070969 | 0.66 |
ENSDART00000020995
|
tmem26b
|
transmembrane protein 26b |
| chr19_-_32600638 | 0.65 |
ENSDART00000143497
|
zgc:91944
|
zgc:91944 |
| chr14_-_32893785 | 0.65 |
ENSDART00000169380
|
slc25a43
|
solute carrier family 25, member 43 |
| chr22_+_7462997 | 0.63 |
ENSDART00000106082
|
zgc:112368
|
zgc:112368 |
| chr19_+_10396042 | 0.63 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr8_-_16697912 | 0.63 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr25_-_18002937 | 0.62 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr10_+_23099890 | 0.62 |
ENSDART00000135890
|
LTN1
|
si:dkey-175g6.5 |
| chr19_-_22843480 | 0.62 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr4_+_7391110 | 0.61 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr13_+_35635672 | 0.61 |
ENSDART00000148481
|
thbs2a
|
thrombospondin 2a |
| chr1_+_34295925 | 0.61 |
ENSDART00000075584
|
kctd12.2
|
potassium channel tetramerisation domain containing 12.2 |
| chr2_+_20866898 | 0.60 |
ENSDART00000150086
|
odr4
|
odr-4 GPCR localization factor homolog |
| chr20_-_54014539 | 0.60 |
ENSDART00000060466
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr8_-_30791266 | 0.59 |
ENSDART00000062220
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr10_+_33895315 | 0.58 |
ENSDART00000142881
|
fryb
|
furry homolog b (Drosophila) |
| chr13_-_45475289 | 0.58 |
ENSDART00000043345
|
rsrp1
|
arginine/serine-rich protein 1 |
| chr15_-_23647078 | 0.57 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr10_-_23099809 | 0.56 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr5_-_34993242 | 0.56 |
ENSDART00000134516
ENSDART00000051295 |
btf3
|
basic transcription factor 3 |
| chr21_-_41588129 | 0.56 |
ENSDART00000164125
|
crebrf
|
creb3 regulatory factor |
| chr14_-_33177935 | 0.56 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr21_-_32081552 | 0.56 |
ENSDART00000135659
|
mat2b
|
methionine adenosyltransferase II, beta |
| chr20_+_30445971 | 0.55 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr22_+_30331414 | 0.54 |
ENSDART00000133482
|
BX649448.3
|
|
| chr11_+_6819050 | 0.54 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr20_+_30578967 | 0.54 |
ENSDART00000010494
|
fgfr1op
|
FGFR1 oncogene partner |
| chr23_+_25354856 | 0.53 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr25_-_16818195 | 0.53 |
ENSDART00000185215
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr8_-_30791089 | 0.53 |
ENSDART00000147332
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr1_-_26063188 | 0.53 |
ENSDART00000168640
|
pdcd4a
|
programmed cell death 4a |
| chr13_-_31647323 | 0.52 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr3_+_5575313 | 0.52 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr10_-_32890617 | 0.52 |
ENSDART00000134922
|
kctd7
|
potassium channel tetramerization domain containing 7 |
| chr10_-_42108137 | 0.51 |
ENSDART00000132976
|
ess2
|
ess-2 splicing factor homolog |
| chr21_-_22398985 | 0.51 |
ENSDART00000138128
|
si:ch73-112l6.1
|
si:ch73-112l6.1 |
| chr7_-_26844064 | 0.51 |
ENSDART00000162241
|
si:ch211-107p11.3
|
si:ch211-107p11.3 |
| chr12_-_22509069 | 0.51 |
ENSDART00000179755
ENSDART00000109707 |
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr25_+_8356707 | 0.51 |
ENSDART00000153708
|
muc5.1
|
mucin 5.1, oligomeric mucus/gel-forming |
| chr8_+_20624510 | 0.50 |
ENSDART00000138604
|
nfic
|
nuclear factor I/C |
| chr16_+_25608778 | 0.50 |
ENSDART00000077484
|
zhx2a
|
zinc fingers and homeoboxes 2a |
| chr20_-_26383368 | 0.49 |
ENSDART00000024518
ENSDART00000087844 |
esr1
|
estrogen receptor 1 |
| chr13_+_22119798 | 0.49 |
ENSDART00000173206
ENSDART00000078652 ENSDART00000165842 |
camk2g2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 2 |
| chr6_-_39167732 | 0.49 |
ENSDART00000153626
|
apof
|
apolipoprotein F |
| chr3_+_18567560 | 0.49 |
ENSDART00000124329
|
cbx8a
|
chromobox homolog 8a (Pc class homolog, Drosophila) |
| chr3_-_7119493 | 0.49 |
ENSDART00000184911
|
BX005085.7
|
|
| chr2_-_42958113 | 0.47 |
ENSDART00000139945
|
oc90
|
otoconin 90 |
| chr5_+_30680804 | 0.46 |
ENSDART00000078049
|
pdzd3b
|
PDZ domain containing 3b |
| chr20_+_10723292 | 0.45 |
ENSDART00000152805
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr17_+_43843536 | 0.45 |
ENSDART00000164097
|
FO704777.1
|
|
| chr3_-_18410968 | 0.45 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr15_-_30816370 | 0.45 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr5_+_36611128 | 0.44 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr21_+_15704556 | 0.44 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr7_-_28658143 | 0.43 |
ENSDART00000173556
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr22_+_18166660 | 0.43 |
ENSDART00000105432
|
borcs8
|
BLOC-1 related complex subunit 8 |
| chr13_-_43149063 | 0.43 |
ENSDART00000099601
|
vsir
|
V-set immunoregulatory receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxj1a+foxj1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.5 | 1.4 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.5 | 1.4 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.4 | 1.7 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 1.0 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.3 | 1.2 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.3 | 0.9 | GO:0030238 | male sex determination(GO:0030238) |
| 0.3 | 1.8 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.3 | 0.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.3 | 0.8 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.3 | 1.0 | GO:0016122 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.2 | 1.4 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.2 | 0.7 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.2 | 2.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.2 | 0.9 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 2.8 | GO:0045824 | negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.2 | 0.5 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 0.9 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 1.2 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.2 | 1.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.3 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.1 | 0.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.9 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.1 | 0.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 1.1 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 2.7 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.1 | 1.3 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 1.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.5 | GO:0006949 | syncytium formation by plasma membrane fusion(GO:0000768) syncytium formation(GO:0006949) |
| 0.1 | 0.3 | GO:0042421 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.5 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.8 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.1 | 0.9 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.1 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 1.0 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 1.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.4 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.2 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 0.8 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.7 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.1 | 1.1 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.9 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 1.0 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.1 | 1.2 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.3 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.1 | 2.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.3 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 1.6 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 0.8 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 2.2 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.2 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.6 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.8 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 1.2 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.3 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.5 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.4 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 1.0 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.8 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 10.5 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 1.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.4 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 1.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.9 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.5 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.6 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0072672 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 1.1 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.5 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.1 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.9 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.7 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.7 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.9 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 1.3 | GO:0019362 | pyridine nucleotide metabolic process(GO:0019362) nicotinamide nucleotide metabolic process(GO:0046496) |
| 0.0 | 0.3 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.2 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.3 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.7 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.4 | GO:0030851 | granulocyte differentiation(GO:0030851) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0001534 | radial spoke(GO:0001534) |
| 0.5 | 2.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 1.0 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.3 | 4.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 2.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 1.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 0.7 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 1.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 0.9 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 1.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.6 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.2 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 2.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.4 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 2.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.2 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.6 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.4 | 1.3 | GO:0047453 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.4 | 1.7 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.4 | 1.8 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.4 | 1.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 1.0 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.3 | 2.3 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.3 | 1.6 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.3 | 1.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.3 | 1.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.3 | 1.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.3 | 1.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 1.0 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.2 | 1.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 1.7 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 0.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 1.5 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.2 | 1.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 2.0 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.2 | 0.7 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 0.5 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.1 | 2.4 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.1 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.4 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 1.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.5 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 1.1 | GO:0070035 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 0.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 0.8 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 2.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.6 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 1.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.4 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.8 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 1.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 1.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.9 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 2.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 2.9 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 2.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 3.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.9 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 3.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 2.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 4.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.1 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |