Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
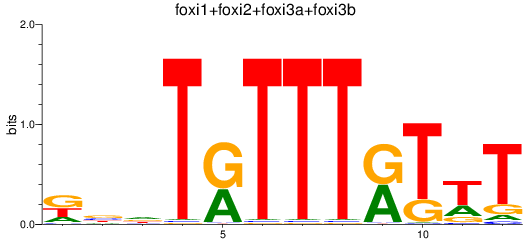
Results for foxi1+foxi2+foxi3a+foxi3b
Z-value: 0.35
Transcription factors associated with foxi1+foxi2+foxi3a+foxi3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxi3b
|
ENSDARG00000009550 | forkhead box I3b |
|
foxi3a
|
ENSDARG00000055926 | forkhead box I3a |
|
foxi2
|
ENSDARG00000069715 | forkhead box I2 |
|
foxi1
|
ENSDARG00000104566 | forkhead box i1 |
|
foxi3b
|
ENSDARG00000112022 | forkhead box I3b |
|
foxi3b
|
ENSDARG00000112303 | forkhead box I3b |
|
foxi2
|
ENSDARG00000116791 | forkhead box I2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxi2 | dr11_v1_chr13_+_13578552_13578552 | 0.68 | 2.0e-03 | Click! |
| foxi3b | dr11_v1_chr14_+_34514336_34514336 | 0.66 | 2.7e-03 | Click! |
| foxi1 | dr11_v1_chr12_-_43664682_43664682 | 0.28 | 2.6e-01 | Click! |
| foxi3a | dr11_v1_chr21_+_30351256_30351256 | 0.08 | 7.4e-01 | Click! |
Activity profile of foxi1+foxi2+foxi3a+foxi3b motif
Sorted Z-values of foxi1+foxi2+foxi3a+foxi3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_26973063 | 0.40 |
ENSDART00000143162
ENSDART00000186210 |
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr10_+_26972755 | 0.39 |
ENSDART00000042162
|
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr19_-_8880688 | 0.37 |
ENSDART00000039629
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr2_-_42492201 | 0.36 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr17_-_11417904 | 0.36 |
ENSDART00000103228
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr17_-_11417551 | 0.35 |
ENSDART00000128291
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr3_-_4501026 | 0.35 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr17_+_25856671 | 0.33 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr11_+_21586335 | 0.33 |
ENSDART00000091182
|
FAM72B
|
zgc:101564 |
| chr19_+_25971000 | 0.31 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr7_-_50367642 | 0.30 |
ENSDART00000134941
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr7_-_13906409 | 0.30 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr11_+_18612421 | 0.28 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr5_-_49951106 | 0.27 |
ENSDART00000135954
|
fam172a
|
family with sequence similarity 172, member A |
| chr3_-_2623176 | 0.27 |
ENSDART00000179792
ENSDART00000123512 |
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr25_-_14087377 | 0.27 |
ENSDART00000124140
|
zgc:101566
|
zgc:101566 |
| chr21_-_26490186 | 0.26 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr22_+_30112390 | 0.26 |
ENSDART00000191715
|
add3a
|
adducin 3 (gamma) a |
| chr15_+_23657051 | 0.26 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr13_-_32626247 | 0.25 |
ENSDART00000100663
|
wdr35
|
WD repeat domain 35 |
| chr2_-_42492445 | 0.25 |
ENSDART00000139929
|
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr5_-_48070779 | 0.25 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr23_-_42810664 | 0.24 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr23_-_24682244 | 0.24 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr1_-_6028876 | 0.24 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr7_-_50367326 | 0.24 |
ENSDART00000141926
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr16_+_18535618 | 0.22 |
ENSDART00000021596
|
rxrbb
|
retinoid x receptor, beta b |
| chr6_+_4255319 | 0.22 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr8_+_23147218 | 0.21 |
ENSDART00000030920
ENSDART00000141175 ENSDART00000146264 |
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr16_+_16265850 | 0.20 |
ENSDART00000181265
|
setd2
|
SET domain containing 2 |
| chr1_-_50611031 | 0.20 |
ENSDART00000148285
|
ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr3_+_53116172 | 0.20 |
ENSDART00000115117
|
brd4
|
bromodomain containing 4 |
| chr10_+_1849874 | 0.20 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr2_+_31437547 | 0.20 |
ENSDART00000141170
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr22_+_23359369 | 0.20 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr5_-_22052852 | 0.18 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr19_+_34274504 | 0.18 |
ENSDART00000132046
|
si:ch211-9n13.3
|
si:ch211-9n13.3 |
| chr5_+_41996889 | 0.17 |
ENSDART00000097580
|
pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr8_+_8643901 | 0.17 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr2_-_57110477 | 0.17 |
ENSDART00000181132
|
slc25a42
|
solute carrier family 25, member 42 |
| chr1_-_39895859 | 0.17 |
ENSDART00000135791
ENSDART00000035739 |
tmem134
|
transmembrane protein 134 |
| chr4_-_12978925 | 0.17 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr18_+_37015185 | 0.17 |
ENSDART00000191305
|
sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr19_-_2822372 | 0.16 |
ENSDART00000109130
ENSDART00000122385 |
recql4
|
RecQ helicase-like 4 |
| chr13_+_37653851 | 0.16 |
ENSDART00000141988
ENSDART00000126902 ENSDART00000100352 |
phf3
|
PHD finger protein 3 |
| chr4_+_6833583 | 0.16 |
ENSDART00000165179
ENSDART00000186134 ENSDART00000174507 |
dock4b
|
dedicator of cytokinesis 4b |
| chr19_-_12965020 | 0.16 |
ENSDART00000128975
|
slc25a32a
|
solute carrier family 25 (mitochondrial folate carrier), member 32a |
| chr23_-_27822920 | 0.16 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr18_-_15532016 | 0.16 |
ENSDART00000165279
|
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr1_+_45839927 | 0.15 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr11_+_42641404 | 0.15 |
ENSDART00000172641
ENSDART00000169938 |
il17rd
|
interleukin 17 receptor D |
| chr1_+_33647682 | 0.15 |
ENSDART00000114384
|
znf654
|
zinc finger protein 654 |
| chr6_-_10037207 | 0.15 |
ENSDART00000179701
|
nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr4_-_12477224 | 0.15 |
ENSDART00000027756
ENSDART00000182706 ENSDART00000127150 |
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr2_+_58841181 | 0.15 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr15_+_38299563 | 0.14 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr16_-_41487589 | 0.14 |
ENSDART00000188115
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr7_+_13830052 | 0.14 |
ENSDART00000191360
|
abhd2a
|
abhydrolase domain containing 2a |
| chr20_-_45709990 | 0.14 |
ENSDART00000027482
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr12_-_31422433 | 0.14 |
ENSDART00000186075
ENSDART00000153172 ENSDART00000066256 |
vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr17_+_32500387 | 0.14 |
ENSDART00000018423
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr15_-_33807758 | 0.14 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr20_-_31252809 | 0.14 |
ENSDART00000137236
|
hpcal1
|
hippocalcin-like 1 |
| chr19_-_9503473 | 0.13 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr16_-_41488023 | 0.13 |
ENSDART00000169312
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr21_+_22558187 | 0.13 |
ENSDART00000167599
|
chek1
|
checkpoint kinase 1 |
| chr7_-_33868903 | 0.13 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr4_+_6833735 | 0.13 |
ENSDART00000136355
|
dock4b
|
dedicator of cytokinesis 4b |
| chr9_-_33785093 | 0.13 |
ENSDART00000140779
ENSDART00000059837 |
fundc1
|
FUN14 domain containing 1 |
| chr1_+_9153141 | 0.13 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr12_+_26467847 | 0.13 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr23_+_33296588 | 0.12 |
ENSDART00000030368
|
si:ch211-226m16.3
|
si:ch211-226m16.3 |
| chr24_-_23716097 | 0.12 |
ENSDART00000084954
ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr15_-_18432673 | 0.12 |
ENSDART00000146853
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr3_+_60589157 | 0.12 |
ENSDART00000165367
|
mettl23
|
methyltransferase like 23 |
| chr2_+_47471647 | 0.12 |
ENSDART00000184199
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr14_+_32926385 | 0.12 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr4_+_13953537 | 0.12 |
ENSDART00000133596
|
pphln1
|
periphilin 1 |
| chr7_+_7511914 | 0.12 |
ENSDART00000172848
|
clcn3
|
chloride channel 3 |
| chr9_+_24088062 | 0.12 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr2_+_47471943 | 0.12 |
ENSDART00000141974
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr18_-_44847855 | 0.12 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr19_-_27462720 | 0.12 |
ENSDART00000135901
|
si:ch73-25f10.6
|
si:ch73-25f10.6 |
| chr19_+_1510971 | 0.11 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr11_+_34523132 | 0.11 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr3_+_20012891 | 0.11 |
ENSDART00000078974
|
tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr1_+_33558555 | 0.11 |
ENSDART00000018472
|
chmp2bb
|
charged multivesicular body protein 2Bb |
| chr12_-_35505610 | 0.11 |
ENSDART00000105518
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr11_+_25693395 | 0.11 |
ENSDART00000110224
|
mon1bb
|
MON1 secretory trafficking family member Bb |
| chr15_+_38299385 | 0.11 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr12_+_8569685 | 0.11 |
ENSDART00000031676
|
nrbf2b
|
nuclear receptor binding factor 2b |
| chr2_-_37797577 | 0.11 |
ENSDART00000110781
|
nfatc4
|
nuclear factor of activated T cells 4 |
| chr3_+_301479 | 0.11 |
ENSDART00000165169
|
CABZ01075611.1
|
|
| chr4_-_20135919 | 0.10 |
ENSDART00000172230
|
cep83
|
centrosomal protein 83 |
| chr4_+_11723852 | 0.10 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr15_+_26941063 | 0.10 |
ENSDART00000149957
|
bcas3
|
breast carcinoma amplified sequence 3 |
| chr5_-_19400166 | 0.10 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr17_+_15788100 | 0.10 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr22_-_10752471 | 0.10 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr1_-_41192059 | 0.10 |
ENSDART00000084665
ENSDART00000135369 |
dok7
|
docking protein 7 |
| chr10_-_41940302 | 0.10 |
ENSDART00000033121
|
morn3
|
MORN repeat containing 3 |
| chr24_-_18659147 | 0.10 |
ENSDART00000166039
|
zgc:66014
|
zgc:66014 |
| chr17_-_29271359 | 0.10 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr17_+_15534815 | 0.10 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr15_-_20933574 | 0.10 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr7_+_21787507 | 0.09 |
ENSDART00000100936
|
tmem88b
|
transmembrane protein 88 b |
| chr18_+_41561285 | 0.09 |
ENSDART00000169621
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr25_+_6451038 | 0.09 |
ENSDART00000009971
|
snx33
|
sorting nexin 33 |
| chr5_-_4931266 | 0.09 |
ENSDART00000067600
|
zbtb43
|
zinc finger and BTB domain containing 43 |
| chr2_-_37043905 | 0.09 |
ENSDART00000056514
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr14_+_34966598 | 0.09 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr8_+_12930216 | 0.09 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr8_+_10862353 | 0.09 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr23_+_22873415 | 0.09 |
ENSDART00000135130
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr24_-_9002038 | 0.09 |
ENSDART00000066783
ENSDART00000150185 |
mppe1
|
metallophosphoesterase 1 |
| chr8_+_16738282 | 0.09 |
ENSDART00000134265
ENSDART00000100698 |
ercc8
|
excision repair cross-complementation group 8 |
| chr19_-_47570672 | 0.09 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr13_+_50778187 | 0.09 |
ENSDART00000159597
|
dntt
|
deoxynucleotidyltransferase, terminal |
| chr19_-_29303788 | 0.09 |
ENSDART00000112167
|
srfbp1
|
serum response factor binding protein 1 |
| chr2_-_37043540 | 0.09 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr23_+_33718602 | 0.09 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr6_-_40922971 | 0.09 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr4_-_20135406 | 0.09 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr15_+_26940569 | 0.09 |
ENSDART00000189636
ENSDART00000077172 |
bcas3
|
breast carcinoma amplified sequence 3 |
| chr12_-_14211067 | 0.08 |
ENSDART00000077903
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr18_+_16192083 | 0.08 |
ENSDART00000133042
|
lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr8_+_25900049 | 0.08 |
ENSDART00000124300
ENSDART00000127618 ENSDART00000024009 |
rhoab
|
ras homolog gene family, member Ab |
| chr16_-_13622794 | 0.08 |
ENSDART00000146953
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr20_-_29498178 | 0.08 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr18_+_41560822 | 0.08 |
ENSDART00000158503
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr17_+_50261603 | 0.08 |
ENSDART00000154503
ENSDART00000154467 |
syncripl
|
synaptotagmin binding, cytoplasmic RNA interacting protein, like |
| chr12_+_25223843 | 0.08 |
ENSDART00000077180
ENSDART00000127454 ENSDART00000122665 |
mta3
|
metastasis associated 1 family, member 3 |
| chr22_-_21755524 | 0.07 |
ENSDART00000149635
|
tle2b
|
transducin like enhancer of split 2b |
| chr8_-_21372446 | 0.07 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr17_+_32622933 | 0.07 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr23_+_33957350 | 0.07 |
ENSDART00000172069
|
snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr5_-_63286077 | 0.07 |
ENSDART00000131274
|
tbcelb
|
tubulin folding cofactor E-like b |
| chr18_-_21725638 | 0.07 |
ENSDART00000089853
|
fan1
|
FANCD2/FANCI-associated nuclease 1 |
| chr3_-_60589292 | 0.07 |
ENSDART00000157822
|
jmjd6
|
jumonji domain containing 6 |
| chr17_+_14965570 | 0.07 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr9_+_34950942 | 0.07 |
ENSDART00000077800
|
tfdp1a
|
transcription factor Dp-1, a |
| chr5_-_33255759 | 0.07 |
ENSDART00000085531
|
prkaa1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr11_-_21586157 | 0.07 |
ENSDART00000190095
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr9_-_7238839 | 0.07 |
ENSDART00000142726
|
creg2
|
cellular repressor of E1A-stimulated genes 2 |
| chr5_-_56924747 | 0.06 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr16_+_5612547 | 0.06 |
ENSDART00000140226
ENSDART00000189352 |
CYTH2
|
si:dkey-283b15.4 |
| chr9_+_27720428 | 0.06 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr22_-_35063526 | 0.06 |
ENSDART00000162211
|
zgc:63733
|
zgc:63733 |
| chr4_-_74892355 | 0.06 |
ENSDART00000188725
|
PHF21B
|
PHD finger protein 21B |
| chr19_-_45650994 | 0.06 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr22_+_10752511 | 0.06 |
ENSDART00000081188
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr17_-_14966384 | 0.06 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr12_-_43428542 | 0.06 |
ENSDART00000192266
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr17_-_2685026 | 0.05 |
ENSDART00000191014
ENSDART00000179309 |
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr22_+_10752787 | 0.05 |
ENSDART00000186542
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_23790748 | 0.05 |
ENSDART00000041877
|
csrnp1a
|
cysteine-serine-rich nuclear protein 1a |
| chr14_+_35024521 | 0.05 |
ENSDART00000158634
ENSDART00000170631 |
ebf3a
|
early B cell factor 3a |
| chr13_-_29505946 | 0.05 |
ENSDART00000026679
|
cdhr1a
|
cadherin-related family member 1a |
| chr10_-_34089779 | 0.05 |
ENSDART00000140070
|
pimr144
|
Pim proto-oncogene, serine/threonine kinase, related 144 |
| chr20_-_14875308 | 0.05 |
ENSDART00000141290
|
dnm3a
|
dynamin 3a |
| chr13_-_29505604 | 0.05 |
ENSDART00000110005
|
cdhr1a
|
cadherin-related family member 1a |
| chr23_-_31372639 | 0.05 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr5_+_61361815 | 0.05 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr7_+_52712807 | 0.05 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr1_+_31674297 | 0.05 |
ENSDART00000044214
|
wbp1lb
|
WW domain binding protein 1-like b |
| chr16_+_38820486 | 0.05 |
ENSDART00000131866
|
trhra
|
thyrotropin-releasing hormone receptor a |
| chr19_+_33476557 | 0.05 |
ENSDART00000181800
|
triqk
|
triple QxxK/R motif containing |
| chr4_-_9609634 | 0.05 |
ENSDART00000067188
|
dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae) |
| chr22_-_28777374 | 0.04 |
ENSDART00000188206
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr7_+_38808027 | 0.04 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr3_-_56871330 | 0.04 |
ENSDART00000014103
|
zgc:112148
|
zgc:112148 |
| chr9_-_41040492 | 0.04 |
ENSDART00000163164
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr2_-_42958619 | 0.04 |
ENSDART00000144317
|
oc90
|
otoconin 90 |
| chr18_+_12147971 | 0.04 |
ENSDART00000162067
ENSDART00000168386 |
fgd4a
|
FYVE, RhoGEF and PH domain containing 4a |
| chr14_+_35023923 | 0.04 |
ENSDART00000172171
|
ebf3a
|
early B cell factor 3a |
| chr5_-_33215261 | 0.04 |
ENSDART00000097935
ENSDART00000134777 |
si:dkey-226m8.10
|
si:dkey-226m8.10 |
| chr10_-_11840353 | 0.04 |
ENSDART00000127581
|
trim23
|
tripartite motif containing 23 |
| chr15_+_42431198 | 0.04 |
ENSDART00000189951
ENSDART00000089694 |
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr2_+_56891858 | 0.04 |
ENSDART00000159912
|
zgc:85843
|
zgc:85843 |
| chr19_-_44091405 | 0.04 |
ENSDART00000132800
|
rad21b
|
RAD21 cohesin complex component b |
| chr16_-_29334672 | 0.04 |
ENSDART00000162835
|
bcan
|
brevican |
| chr10_-_43568239 | 0.03 |
ENSDART00000131731
ENSDART00000097433 ENSDART00000131309 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr1_-_9114936 | 0.03 |
ENSDART00000163624
ENSDART00000132910 |
si:ch211-14k19.8
vap
|
si:ch211-14k19.8 vascular associated protein |
| chr24_-_4765740 | 0.03 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr22_+_20720808 | 0.03 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr5_+_60919378 | 0.03 |
ENSDART00000184915
|
doc2b
|
double C2-like domains, beta |
| chr5_+_37837245 | 0.03 |
ENSDART00000171617
|
epd
|
ependymin |
| chr22_-_28777557 | 0.03 |
ENSDART00000135214
ENSDART00000131761 ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr17_-_33415740 | 0.03 |
ENSDART00000135218
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr10_-_24343507 | 0.03 |
ENSDART00000002974
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr18_-_20560007 | 0.03 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr5_+_42092227 | 0.03 |
ENSDART00000097583
ENSDART00000171678 |
ubb
|
ubiquitin B |
| chr19_+_16064439 | 0.03 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr13_+_27316934 | 0.03 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr7_+_42206847 | 0.03 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr18_-_25855263 | 0.03 |
ENSDART00000042074
|
sema4ba
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ba |
| chr17_-_1705013 | 0.03 |
ENSDART00000182864
|
CABZ01086293.1
|
|
| chr7_+_38510197 | 0.03 |
ENSDART00000173468
ENSDART00000100479 |
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr6_-_35446110 | 0.02 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr3_+_45778810 | 0.02 |
ENSDART00000003338
ENSDART00000155409 |
unkl
|
unkempt family zinc finger-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxi1+foxi2+foxi3a+foxi3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.2 | GO:1905067 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.0 | 0.5 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.6 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.1 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.2 | GO:0006203 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.0 | GO:0061178 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.1 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.0 | GO:0036076 | ligamentous ossification(GO:0036076) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.2 | GO:0015230 | folic acid transporter activity(GO:0008517) FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.2 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.0 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |