Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
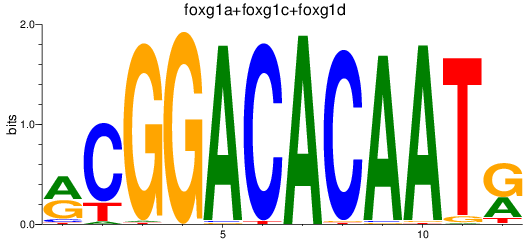
Results for foxg1a+foxg1c+foxg1d
Z-value: 0.34
Transcription factors associated with foxg1a+foxg1c+foxg1d
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxg1c
|
ENSDARG00000068380 | forkhead box G1c |
|
foxg1d
|
ENSDARG00000070053 | forkhead box G1d |
|
foxg1a
|
ENSDARG00000070769 | forkhead box G1a |
|
foxg1c
|
ENSDARG00000114414 | forkhead box G1c |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxg1a | dr11_v1_chr17_-_29119362_29119362 | -0.45 | 6.0e-02 | Click! |
| foxg1c | dr11_v1_chr18_-_40901707_40901707 | -0.39 | 1.1e-01 | Click! |
| foxg1d | dr11_v1_chr13_+_255067_255067 | -0.32 | 2.0e-01 | Click! |
Activity profile of foxg1a+foxg1c+foxg1d motif
Sorted Z-values of foxg1a+foxg1c+foxg1d motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_4252221 | 0.56 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr5_+_37720777 | 0.54 |
ENSDART00000076083
|
cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr3_+_28860283 | 0.41 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr24_+_18286427 | 0.39 |
ENSDART00000055443
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr3_-_31069776 | 0.34 |
ENSDART00000167462
|
elob
|
elongin B |
| chr2_-_1486023 | 0.32 |
ENSDART00000113774
|
si:ch211-188c16.1
|
si:ch211-188c16.1 |
| chr18_-_34170918 | 0.31 |
ENSDART00000015079
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr18_+_20482369 | 0.30 |
ENSDART00000100668
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr18_-_34171280 | 0.29 |
ENSDART00000122321
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr20_-_14665002 | 0.29 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr24_-_37640705 | 0.27 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr19_-_8096984 | 0.26 |
ENSDART00000146987
|
si:dkey-266f7.9
|
si:dkey-266f7.9 |
| chr1_-_46875493 | 0.25 |
ENSDART00000115081
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr4_+_14343706 | 0.16 |
ENSDART00000142845
|
prl2
|
prolactin 2 |
| chr24_+_37640626 | 0.16 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr8_+_19621511 | 0.16 |
ENSDART00000017128
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr7_+_9189547 | 0.15 |
ENSDART00000169783
|
pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr18_+_18104235 | 0.15 |
ENSDART00000145342
|
cbln1
|
cerebellin 1 precursor |
| chr8_+_19621731 | 0.14 |
ENSDART00000144667
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr1_-_50710468 | 0.14 |
ENSDART00000080389
|
fam13a
|
family with sequence similarity 13, member A |
| chr5_+_22791686 | 0.14 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr15_-_16155729 | 0.13 |
ENSDART00000192212
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr19_-_11949996 | 0.12 |
ENSDART00000163478
ENSDART00000167299 |
cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chr11_+_13058613 | 0.12 |
ENSDART00000161532
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr16_-_33806390 | 0.11 |
ENSDART00000160671
|
rspo1
|
R-spondin 1 |
| chr5_+_60934534 | 0.10 |
ENSDART00000065025
|
rph3al
|
rabphilin 3A-like (without C2 domains) |
| chr17_-_11439815 | 0.09 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr10_+_29850330 | 0.09 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr15_-_8763106 | 0.08 |
ENSDART00000154171
|
arhgap35a
|
Rho GTPase activating protein 35a |
| chr10_+_29849977 | 0.08 |
ENSDART00000180242
|
hspa8
|
heat shock protein 8 |
| chr3_+_23248542 | 0.08 |
ENSDART00000185765
ENSDART00000192332 |
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr19_-_19025998 | 0.08 |
ENSDART00000186156
ENSDART00000163359 ENSDART00000167951 |
dync1li1
|
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr20_-_54564018 | 0.08 |
ENSDART00000099832
|
zgc:153012
|
zgc:153012 |
| chr19_-_22541284 | 0.08 |
ENSDART00000019505
|
pleca
|
plectin a |
| chr24_+_19591893 | 0.07 |
ENSDART00000152026
|
slco5a1a
|
solute carrier organic anion transporter family member 5A1a |
| chr4_+_3980247 | 0.07 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr11_-_42739148 | 0.06 |
ENSDART00000183584
|
CU459094.2
|
|
| chr2_-_7246338 | 0.06 |
ENSDART00000186735
|
zgc:153115
|
zgc:153115 |
| chr18_-_44935174 | 0.05 |
ENSDART00000081025
|
pex16
|
peroxisomal biogenesis factor 16 |
| chr16_-_18702249 | 0.05 |
ENSDART00000191595
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr24_-_36316104 | 0.05 |
ENSDART00000048046
|
naglu
|
N-acetylglucosaminidase, alpha |
| chr14_+_12316581 | 0.05 |
ENSDART00000170115
ENSDART00000149757 |
si:ch211-125c23.3
cx31.7
|
si:ch211-125c23.3 connexin 31.7 |
| chr17_-_15657029 | 0.04 |
ENSDART00000153925
|
fut9a
|
fucosyltransferase 9a |
| chr25_-_16768078 | 0.04 |
ENSDART00000025186
|
galnt8a.1
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 1 |
| chr12_+_39685485 | 0.04 |
ENSDART00000163403
|
LO017650.1
|
|
| chr20_+_34320635 | 0.04 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr4_-_8902406 | 0.04 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr2_+_30721466 | 0.03 |
ENSDART00000128982
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr2_+_35880600 | 0.03 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr14_-_2361692 | 0.03 |
ENSDART00000167696
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.4 |
| chr15_-_30816370 | 0.03 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr11_+_583725 | 0.03 |
ENSDART00000189415
|
mkrn2os.2
|
MKRN2 opposite strand, tandem duplicate 2 |
| chr11_-_26666501 | 0.02 |
ENSDART00000188067
ENSDART00000111539 |
efcc1
|
EF-hand and coiled-coil domain containing 1 |
| chr1_+_59090583 | 0.02 |
ENSDART00000150658
|
mfap4
|
microfibril associated protein 4 |
| chr7_-_51953613 | 0.02 |
ENSDART00000142042
|
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr2_-_59345920 | 0.02 |
ENSDART00000134662
|
ftr37
|
finTRIM family, member 37 |
| chr18_-_5509616 | 0.02 |
ENSDART00000142945
|
bloc1s6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr7_-_51953807 | 0.02 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr1_+_59090743 | 0.02 |
ENSDART00000100199
|
mfap4
|
microfibril associated protein 4 |
| chr25_+_21267579 | 0.02 |
ENSDART00000152015
|
fgl2b
|
fibrinogen-like 2b |
| chr15_-_23522653 | 0.01 |
ENSDART00000144685
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr4_+_1283068 | 0.01 |
ENSDART00000167233
|
chrm2a
|
cholinergic receptor, muscarinic 2a |
| chr17_+_31719034 | 0.01 |
ENSDART00000077077
|
nubpl
|
nucleotide binding protein-like |
| chr2_+_30721070 | 0.01 |
ENSDART00000099052
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr2_-_59231017 | 0.00 |
ENSDART00000143980
ENSDART00000184190 |
ftr29
|
finTRIM family, member 29 |
| chr3_-_4591643 | 0.00 |
ENSDART00000138144
|
ftr50
|
finTRIM family, member 50 |
| chr24_-_6038025 | 0.00 |
ENSDART00000077819
ENSDART00000139216 |
ftr61
|
finTRIM family, member 61 |
| chr25_-_16742438 | 0.00 |
ENSDART00000156091
|
galnt8a.1
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 1 |
| chr8_+_25616946 | 0.00 |
ENSDART00000133983
|
slc38a5a
|
solute carrier family 38, member 5a |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxg1a+foxg1c+foxg1d
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.1 | 0.6 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.5 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.3 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.6 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.1 | 0.6 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.3 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |