Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for foxa_foxa1_foxa2
Z-value: 0.65
Transcription factors associated with foxa_foxa1_foxa2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
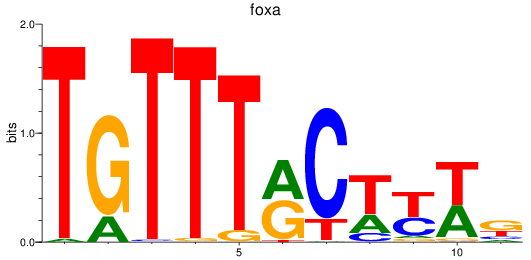
foxa
|
ENSDARG00000087094 | forkhead box A sequence |
|
foxa
|
ENSDARG00000110743 | forkhead box A sequence |
|
foxa
|
ENSDARG00000115019 | forkhead box A sequence |
|
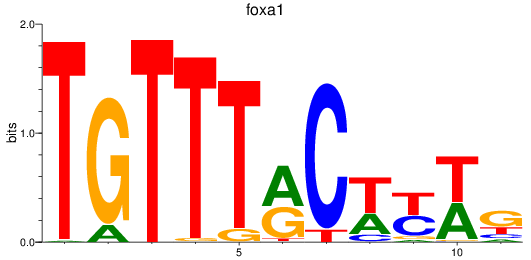
foxa1
|
ENSDARG00000102138 | forkhead box A1 |
|
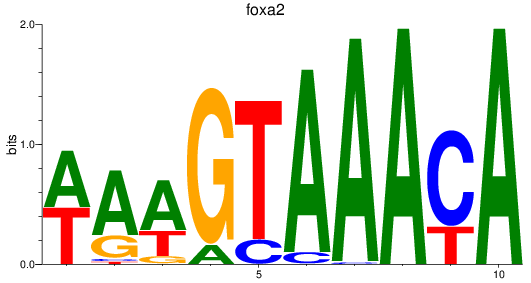
foxa2
|
ENSDARG00000003411 | forkhead box A2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxa2 | dr11_v1_chr17_-_42568498_42568498 | -0.96 | 5.5e-10 | Click! |
| foxa1 | dr11_v1_chr17_+_10318071_10318071 | -0.94 | 9.2e-09 | Click! |
| foxa | dr11_v1_chr14_-_33981544_33981544 | 0.84 | 1.2e-05 | Click! |
Activity profile of foxa_foxa1_foxa2 motif
Sorted Z-values of foxa_foxa1_foxa2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_38299385 | 2.57 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr19_-_43757568 | 2.53 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr1_+_45707219 | 2.48 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr23_-_22523303 | 2.46 |
ENSDART00000079019
|
spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr15_+_38299563 | 2.40 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr22_+_24559947 | 2.00 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr14_+_15155684 | 2.00 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr16_-_26820634 | 1.92 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr17_+_24036791 | 1.91 |
ENSDART00000140767
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr10_+_7709724 | 1.78 |
ENSDART00000097670
|
ggcx
|
gamma-glutamyl carboxylase |
| chr17_+_32623931 | 1.73 |
ENSDART00000144217
|
ctsba
|
cathepsin Ba |
| chr7_+_67699178 | 1.69 |
ENSDART00000160086
|
zgc:162592
|
zgc:162592 |
| chr13_-_32726178 | 1.56 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr12_-_48188928 | 1.49 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr17_-_30652738 | 1.46 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr23_-_36449111 | 1.45 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr9_-_6991650 | 1.44 |
ENSDART00000081718
|
slc9a2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr19_+_7549854 | 1.42 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr9_+_33154841 | 1.42 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr12_+_22404108 | 1.40 |
ENSDART00000153055
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr24_+_19210001 | 1.33 |
ENSDART00000179373
ENSDART00000139299 |
zgc:162928
|
zgc:162928 |
| chr16_-_41714988 | 1.32 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr17_-_15188440 | 1.28 |
ENSDART00000151885
|
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr8_-_25771474 | 1.25 |
ENSDART00000193883
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr20_+_46741074 | 1.24 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr1_+_45969240 | 1.24 |
ENSDART00000042086
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr11_-_35171162 | 1.23 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr5_+_41496490 | 1.22 |
ENSDART00000039369
|
fancg
|
Fanconi anemia, complementation group G |
| chr3_+_40255408 | 1.21 |
ENSDART00000074746
|
smcr8a
|
Smith-Magenis syndrome chromosome region, candidate 8a |
| chr3_+_46762703 | 1.17 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr22_+_105376 | 1.15 |
ENSDART00000059140
|
slc25a20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr5_-_33236637 | 1.15 |
ENSDART00000085512
ENSDART00000144694 |
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr19_-_46091497 | 1.13 |
ENSDART00000178772
ENSDART00000167255 |
ptdss1b
si:dkey-108k24.2
|
phosphatidylserine synthase 1b si:dkey-108k24.2 |
| chr18_-_44935174 | 1.09 |
ENSDART00000081025
|
pex16
|
peroxisomal biogenesis factor 16 |
| chr13_-_23956178 | 1.05 |
ENSDART00000133646
|
phactr2
|
phosphatase and actin regulator 2 |
| chr16_+_38940758 | 1.04 |
ENSDART00000102482
ENSDART00000136215 |
eny2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr7_+_52712807 | 1.04 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr8_-_30338872 | 1.04 |
ENSDART00000137583
|
dock8
|
dedicator of cytokinesis 8 |
| chr12_-_48199227 | 1.02 |
ENSDART00000171058
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr8_-_1219815 | 1.01 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr20_-_52928541 | 1.00 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr13_-_23956361 | 0.99 |
ENSDART00000101150
|
phactr2
|
phosphatase and actin regulator 2 |
| chr18_-_13121983 | 0.97 |
ENSDART00000092648
|
rxylt1
|
ribitol xylosyltransferase 1 |
| chr5_-_31772559 | 0.97 |
ENSDART00000183879
|
fam102ab
|
family with sequence similarity 102, member Ab |
| chr14_-_6931889 | 0.97 |
ENSDART00000166439
|
si:ch211-266k2.1
|
si:ch211-266k2.1 |
| chr7_-_26601307 | 0.97 |
ENSDART00000188934
|
plscr3b
|
phospholipid scramblase 3b |
| chr11_+_11120532 | 0.97 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr9_+_50110763 | 0.96 |
ENSDART00000162990
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr9_+_42607138 | 0.96 |
ENSDART00000138133
ENSDART00000002027 |
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr20_-_28361574 | 0.96 |
ENSDART00000103352
|
vps18
|
vacuolar protein sorting 18 homolog (S. cerevisiae) |
| chr6_-_40657653 | 0.96 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr16_-_35532937 | 0.95 |
ENSDART00000193209
|
ctps1b
|
CTP synthase 1b |
| chr20_+_30378803 | 0.95 |
ENSDART00000148242
ENSDART00000169140 ENSDART00000062441 |
rnaseh1
|
ribonuclease H1 |
| chr16_-_25233515 | 0.92 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr21_+_520479 | 0.92 |
ENSDART00000168054
|
CABZ01079760.1
|
|
| chr14_-_16082806 | 0.92 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr6_+_4371137 | 0.92 |
ENSDART00000168973
|
rbm26
|
RNA binding motif protein 26 |
| chr9_-_46072805 | 0.91 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr2_+_1988036 | 0.91 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr13_-_33114933 | 0.91 |
ENSDART00000140543
ENSDART00000075953 |
zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr5_-_31773208 | 0.87 |
ENSDART00000137556
ENSDART00000122066 |
fam102ab
|
family with sequence similarity 102, member Ab |
| chr9_+_41459759 | 0.86 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr20_+_29209767 | 0.85 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr6_+_4370935 | 0.85 |
ENSDART00000192368
|
rbm26
|
RNA binding motif protein 26 |
| chr14_+_32918484 | 0.85 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr21_+_20386865 | 0.84 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr1_-_9277986 | 0.83 |
ENSDART00000146065
ENSDART00000114876 ENSDART00000132812 |
ubn1
|
ubinuclein 1 |
| chr8_-_35960987 | 0.82 |
ENSDART00000160503
|
slc15a4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr23_-_25686894 | 0.81 |
ENSDART00000181420
ENSDART00000088208 |
lrp1ab
|
low density lipoprotein receptor-related protein 1Ab |
| chr23_-_10722664 | 0.80 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr20_+_29209926 | 0.80 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr18_-_22094102 | 0.80 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr6_+_38626926 | 0.79 |
ENSDART00000190339
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr5_-_23675222 | 0.79 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr3_-_55525627 | 0.79 |
ENSDART00000189234
|
tex2
|
testis expressed 2 |
| chr6_-_3998199 | 0.79 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr1_-_354115 | 0.79 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr8_-_42712573 | 0.78 |
ENSDART00000132229
ENSDART00000137258 |
si:ch73-138n13.1
|
si:ch73-138n13.1 |
| chr4_-_16824556 | 0.78 |
ENSDART00000165289
ENSDART00000185839 |
gys2
|
glycogen synthase 2 |
| chr6_+_40952031 | 0.78 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr14_+_7699443 | 0.76 |
ENSDART00000123139
|
brd8
|
bromodomain containing 8 |
| chr4_-_16824231 | 0.76 |
ENSDART00000014007
|
gys2
|
glycogen synthase 2 |
| chr6_-_10728057 | 0.76 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr6_+_38626684 | 0.76 |
ENSDART00000086533
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr5_+_28271412 | 0.76 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr10_+_11260170 | 0.74 |
ENSDART00000155742
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr19_-_25119443 | 0.73 |
ENSDART00000148953
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr3_-_34528306 | 0.73 |
ENSDART00000023039
|
sept9a
|
septin 9a |
| chr20_+_29209615 | 0.73 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr20_-_25902141 | 0.73 |
ENSDART00000142611
ENSDART00000024821 |
elmsan1a
|
ELM2 and Myb/SANT-like domain containing 1a |
| chr5_+_67835595 | 0.72 |
ENSDART00000159386
|
si:ch211-271b14.1
|
si:ch211-271b14.1 |
| chr15_-_25365570 | 0.72 |
ENSDART00000152754
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr2_-_32262287 | 0.72 |
ENSDART00000056621
ENSDART00000039717 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr22_+_1940595 | 0.72 |
ENSDART00000163506
|
znf1167
|
zinc finger protein 1167 |
| chr17_+_33415319 | 0.71 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr3_+_54168007 | 0.71 |
ENSDART00000109894
|
olfm2a
|
olfactomedin 2a |
| chr4_-_10835620 | 0.71 |
ENSDART00000150739
|
ppfibp1a
|
PTPRF interacting protein, binding protein 1a (liprin beta 1) |
| chr15_+_40079468 | 0.70 |
ENSDART00000154947
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr15_-_25365319 | 0.70 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr14_+_50918769 | 0.70 |
ENSDART00000146918
|
rnf44
|
ring finger protein 44 |
| chr14_+_34495216 | 0.70 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr10_-_36633882 | 0.69 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr7_-_58098814 | 0.69 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr3_+_19685873 | 0.67 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr9_+_24065855 | 0.67 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr12_-_28537615 | 0.66 |
ENSDART00000067762
|
MYO1D
|
si:ch211-94l19.4 |
| chr20_-_28800999 | 0.65 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr17_-_26719821 | 0.64 |
ENSDART00000186813
|
calm1a
|
calmodulin 1a |
| chr11_-_11336986 | 0.64 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr16_-_44673851 | 0.63 |
ENSDART00000015139
|
dcaf13
|
ddb1 and cul4 associated factor 13 |
| chr5_-_66749535 | 0.62 |
ENSDART00000132183
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr6_+_6780873 | 0.62 |
ENSDART00000011865
|
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr7_-_29723761 | 0.62 |
ENSDART00000173560
|
vps13c
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr24_+_19542323 | 0.62 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr2_-_21167652 | 0.61 |
ENSDART00000185792
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr19_+_25971000 | 0.60 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr22_-_11520405 | 0.60 |
ENSDART00000063157
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr11_-_14102131 | 0.60 |
ENSDART00000085158
ENSDART00000191962 |
tmem259
|
transmembrane protein 259 |
| chr5_+_36650096 | 0.60 |
ENSDART00000111414
|
alkbh6
|
alkB homolog 6 |
| chr8_-_16725959 | 0.60 |
ENSDART00000183593
|
depdc1a
|
DEP domain containing 1a |
| chr1_+_51537250 | 0.60 |
ENSDART00000152789
|
etaa1
|
ETAA1, ATR kinase activator |
| chr3_+_32663865 | 0.59 |
ENSDART00000190240
|
si:dkey-16l2.16
|
si:dkey-16l2.16 |
| chr21_-_30030644 | 0.59 |
ENSDART00000190810
|
CU855895.2
|
|
| chr17_+_33415542 | 0.59 |
ENSDART00000183169
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr10_-_15128771 | 0.58 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr17_-_14966384 | 0.57 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr7_+_36898850 | 0.57 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr23_-_10723009 | 0.57 |
ENSDART00000189721
|
foxp1a
|
forkhead box P1a |
| chr16_-_17713859 | 0.57 |
ENSDART00000149275
|
zgc:174935
|
zgc:174935 |
| chr19_+_7636941 | 0.56 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr3_-_21280373 | 0.56 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr20_+_23742574 | 0.56 |
ENSDART00000050559
|
sh3rf1
|
SH3 domain containing ring finger 1 |
| chr12_+_45238292 | 0.55 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr19_-_7272921 | 0.55 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr13_-_33207367 | 0.53 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr10_+_11261576 | 0.53 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr22_+_33135253 | 0.52 |
ENSDART00000004504
|
dag1
|
dystroglycan 1 |
| chr2_-_4032732 | 0.52 |
ENSDART00000158335
|
rab18b
|
RAB18B, member RAS oncogene family |
| chr16_+_31511739 | 0.52 |
ENSDART00000049420
|
ndrg1b
|
N-myc downstream regulated 1b |
| chr21_+_26522571 | 0.51 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr1_-_8917902 | 0.51 |
ENSDART00000137900
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr5_+_31214341 | 0.51 |
ENSDART00000133432
|
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr5_+_52625975 | 0.51 |
ENSDART00000170341
ENSDART00000168317 |
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr17_-_4252221 | 0.51 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr21_+_19547806 | 0.50 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr2_-_57227542 | 0.49 |
ENSDART00000182675
ENSDART00000159480 |
btbd2b
|
BTB (POZ) domain containing 2b |
| chr6_-_41138854 | 0.49 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr10_-_14929392 | 0.49 |
ENSDART00000137430
|
smad2
|
SMAD family member 2 |
| chr23_-_3759345 | 0.49 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr11_+_18612421 | 0.48 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr12_-_22238004 | 0.48 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr22_+_39007533 | 0.48 |
ENSDART00000185958
ENSDART00000129848 |
fam208aa
|
family with sequence similarity 208, member Aa |
| chr24_-_17389263 | 0.47 |
ENSDART00000122757
|
cul1b
|
cullin 1b |
| chr17_-_37474689 | 0.47 |
ENSDART00000103980
|
crip2
|
cysteine-rich protein 2 |
| chr6_-_40922971 | 0.47 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr17_+_25213229 | 0.47 |
ENSDART00000110451
|
arhgef10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr10_-_17710637 | 0.46 |
ENSDART00000147553
|
stard7
|
StAR-related lipid transfer (START) domain containing 7 |
| chr17_+_32158951 | 0.46 |
ENSDART00000165348
ENSDART00000108736 |
adam12
|
ADAM metallopeptidase domain 12 |
| chr3_-_26787430 | 0.45 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr22_+_1947494 | 0.45 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr11_+_35171406 | 0.45 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr24_-_21172122 | 0.45 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr13_+_37653851 | 0.45 |
ENSDART00000141988
ENSDART00000126902 ENSDART00000100352 |
phf3
|
PHD finger protein 3 |
| chr12_-_28794957 | 0.45 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr15_+_21712328 | 0.44 |
ENSDART00000192553
|
NKAPD1
|
zgc:162339 |
| chr17_-_21162821 | 0.44 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr23_-_27501923 | 0.44 |
ENSDART00000188394
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr15_-_28908027 | 0.44 |
ENSDART00000182790
ENSDART00000192461 |
eml2
|
echinoderm microtubule associated protein like 2 |
| chr6_-_41135215 | 0.44 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr4_-_9579299 | 0.43 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr20_-_33487729 | 0.43 |
ENSDART00000061843
|
smek1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr17_+_32622933 | 0.42 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr6_+_49723289 | 0.42 |
ENSDART00000190452
|
stx16
|
syntaxin 16 |
| chr1_+_11107688 | 0.41 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr15_-_47338769 | 0.41 |
ENSDART00000164983
ENSDART00000176289 |
anapc15
|
anaphase promoting complex subunit 15 |
| chr9_-_44905867 | 0.40 |
ENSDART00000138316
ENSDART00000131252 ENSDART00000179383 ENSDART00000159337 |
zgc:66484
|
zgc:66484 |
| chr17_-_21280185 | 0.39 |
ENSDART00000123198
|
hspa12a
|
heat shock protein 12A |
| chr12_+_23991639 | 0.39 |
ENSDART00000003143
|
psme4b
|
proteasome activator subunit 4b |
| chr14_+_32926385 | 0.39 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr19_+_32807236 | 0.38 |
ENSDART00000008698
|
stk3
|
serine/threonine kinase 3 (STE20 homolog, yeast) |
| chr12_+_35203091 | 0.38 |
ENSDART00000153022
|
ndst2b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2b |
| chr25_+_28253844 | 0.38 |
ENSDART00000151891
|
cadps2
|
Ca++-dependent secretion activator 2 |
| chr23_+_39346774 | 0.37 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr21_-_26028205 | 0.37 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr2_-_30200206 | 0.37 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr8_+_16676894 | 0.37 |
ENSDART00000076586
|
si:ch211-198n5.11
|
si:ch211-198n5.11 |
| chr17_+_44030692 | 0.37 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr4_-_13156971 | 0.36 |
ENSDART00000182164
|
grip1
|
glutamate receptor interacting protein 1 |
| chr16_+_47207691 | 0.36 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr6_+_51932267 | 0.36 |
ENSDART00000156256
|
angpt4
|
angiopoietin 4 |
| chr16_+_19029297 | 0.36 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr18_-_39702327 | 0.35 |
ENSDART00000149158
|
dmxl2
|
Dmx-like 2 |
| chr20_+_34502606 | 0.35 |
ENSDART00000139739
|
gorab
|
golgin, rab6-interacting |
| chr4_+_14981854 | 0.35 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr23_-_31763753 | 0.35 |
ENSDART00000053399
|
aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr4_+_1619584 | 0.34 |
ENSDART00000148486
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr13_+_29925397 | 0.34 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr7_+_38808027 | 0.34 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr15_-_5624361 | 0.34 |
ENSDART00000176446
ENSDART00000114410 |
wdr62
|
WD repeat domain 62 |
| chr1_-_28861226 | 0.33 |
ENSDART00000075502
|
timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxa_foxa1_foxa2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.4 | 1.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.3 | 1.8 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.3 | 0.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 1.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 0.7 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.2 | 0.9 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.2 | 2.8 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 1.4 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 0.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 1.0 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.2 | 1.4 | GO:0042539 | hypotonic response(GO:0006971) hypotonic salinity response(GO:0042539) |
| 0.2 | 0.9 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 0.8 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.7 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.9 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.4 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.6 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.8 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.0 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 0.4 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.5 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.6 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.4 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.1 | 1.0 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 1.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.6 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.1 | 0.3 | GO:0034036 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.5 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.6 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.6 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.8 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 1.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.4 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.6 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.3 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.1 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 1.6 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.2 | GO:0045191 | regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.1 | 0.6 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.4 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.5 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 0.2 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 | 0.5 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 1.6 | GO:0009749 | response to glucose(GO:0009749) |
| 0.1 | 0.4 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.4 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.3 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 0.5 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 1.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.3 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.1 | 0.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.7 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.5 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.5 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 1.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 1.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.4 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.8 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.0 | 1.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.4 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.2 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 1.1 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.3 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.1 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.0 | 0.3 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.2 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.0 | 0.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.7 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.4 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.9 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.6 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0032392 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.0 | 1.3 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 2.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:2000252 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.3 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.2 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.5 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.1 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.0 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0048942 | myelination of lateral line nerve axons(GO:0048897) posterior lateral line nerve glial cell differentiation(GO:0048931) myelination of posterior lateral line nerve axons(GO:0048932) lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048938) posterior lateral line nerve glial cell development(GO:0048941) posterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048942) |
| 0.0 | 0.8 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 1.4 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.1 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.5 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.2 | 1.0 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 0.7 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 0.7 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 1.0 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 1.0 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 1.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.2 | GO:0044279 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 2.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 0.5 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 0.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 3.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.6 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 1.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 5.7 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.7 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 1.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.6 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.3 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.0 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.4 | 1.8 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.4 | 1.5 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.3 | 1.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 1.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 1.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 3.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.6 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.4 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.3 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.9 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.3 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.6 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.3 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0070883 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.0 | 0.3 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.0 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.6 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.2 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 1.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 4.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.0 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.0 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.0 | GO:0070643 | vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.0 | GO:0051380 | norepinephrine binding(GO:0051380) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 0.5 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 1.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |