Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
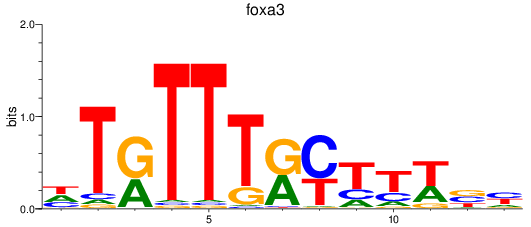
Results for foxa3
Z-value: 0.51
Transcription factors associated with foxa3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxa3
|
ENSDARG00000012788 | forkhead box A3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxa3 | dr11_v1_chr18_-_46354269_46354269 | 0.79 | 9.7e-05 | Click! |
Activity profile of foxa3 motif
Sorted Z-values of foxa3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_52411530 | 1.78 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr16_+_23978978 | 1.63 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr6_-_40352215 | 1.51 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr18_+_7456888 | 1.18 |
ENSDART00000081468
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr12_-_2993095 | 1.16 |
ENSDART00000152316
|
si:dkey-202c14.3
|
si:dkey-202c14.3 |
| chr1_-_54622227 | 1.16 |
ENSDART00000049010
|
tekt4
|
tektin 4 |
| chr3_-_43770876 | 1.14 |
ENSDART00000160162
|
zgc:92162
|
zgc:92162 |
| chr4_-_9722568 | 1.13 |
ENSDART00000067190
|
tspan9b
|
tetraspanin 9b |
| chr1_-_10806625 | 1.11 |
ENSDART00000139749
|
si:ch73-222h13.1
|
si:ch73-222h13.1 |
| chr14_-_8724290 | 1.10 |
ENSDART00000161171
|
pimr56
|
Pim proto-oncogene, serine/threonine kinase, related 56 |
| chr23_+_42813415 | 1.08 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr16_-_54455573 | 1.03 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr12_+_26467847 | 1.03 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr20_-_43462190 | 1.00 |
ENSDART00000100697
|
pimr133
|
Pim proto-oncogene, serine/threonine kinase, related 133 |
| chr20_-_1383916 | 0.92 |
ENSDART00000152373
|
scara5
|
scavenger receptor class A, member 5 (putative) |
| chr12_-_23129453 | 0.89 |
ENSDART00000077453
|
armc4
|
armadillo repeat containing 4 |
| chr4_-_12795030 | 0.85 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr8_+_25902170 | 0.82 |
ENSDART00000193130
|
rhoab
|
ras homolog gene family, member Ab |
| chr7_+_38395197 | 0.82 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr21_-_21324828 | 0.81 |
ENSDART00000136033
|
si:ch211-191j22.7
|
si:ch211-191j22.7 |
| chr9_+_44431174 | 0.78 |
ENSDART00000149726
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr18_+_20566817 | 0.78 |
ENSDART00000100716
|
bida
|
BH3 interacting domain death agonist |
| chr4_-_13921185 | 0.78 |
ENSDART00000143202
ENSDART00000080334 |
yaf2
|
YY1 associated factor 2 |
| chr8_+_24747865 | 0.76 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr7_-_33023404 | 0.76 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr14_-_36799280 | 0.76 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr14_-_25040359 | 0.76 |
ENSDART00000162556
|
TMEM216
|
si:rp71-1d10.5 |
| chr14_-_17576391 | 0.75 |
ENSDART00000161355
ENSDART00000168959 |
rnf4
|
ring finger protein 4 |
| chr7_-_58729894 | 0.73 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr14_-_24397718 | 0.73 |
ENSDART00000110164
|
si:ch211-163m17.4
|
si:ch211-163m17.4 |
| chr1_-_45752010 | 0.72 |
ENSDART00000101410
|
si:ch211-214c7.5
|
si:ch211-214c7.5 |
| chr20_+_52546186 | 0.72 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr3_+_24986145 | 0.70 |
ENSDART00000055428
|
cbx7a
|
chromobox homolog 7a |
| chr20_-_14925281 | 0.70 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr4_-_12795436 | 0.68 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr11_+_35364445 | 0.67 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr21_-_17296789 | 0.67 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr15_+_36941490 | 0.67 |
ENSDART00000172664
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr4_-_149334 | 0.66 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr6_+_24817852 | 0.66 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr25_-_13188214 | 0.65 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr1_+_26071869 | 0.65 |
ENSDART00000059264
|
mxd4
|
MAX dimerization protein 4 |
| chr5_-_16351306 | 0.64 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr4_+_12358822 | 0.64 |
ENSDART00000172557
ENSDART00000150627 |
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr3_-_45298487 | 0.63 |
ENSDART00000102245
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr16_+_23961276 | 0.63 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr14_-_4120636 | 0.63 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr5_+_49744713 | 0.61 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr20_-_29474859 | 0.61 |
ENSDART00000152906
ENSDART00000045249 |
scg5
|
secretogranin V |
| chr1_-_45752261 | 0.61 |
ENSDART00000141756
|
si:ch211-214c7.5
|
si:ch211-214c7.5 |
| chr20_-_29475172 | 0.61 |
ENSDART00000183164
|
scg5
|
secretogranin V |
| chr1_+_38362412 | 0.60 |
ENSDART00000075086
|
cep44
|
centrosomal protein 44 |
| chr16_-_21785261 | 0.59 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr5_-_32882162 | 0.59 |
ENSDART00000085769
|
lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr4_+_14926948 | 0.59 |
ENSDART00000019647
|
psmc2
|
proteasome 26S subunit, ATPase 2 |
| chr15_+_46386261 | 0.58 |
ENSDART00000191793
|
igsf11
|
immunoglobulin superfamily member 11 |
| chr22_-_10110959 | 0.58 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr25_+_19149241 | 0.58 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr7_+_52766211 | 0.57 |
ENSDART00000186191
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr16_-_50203058 | 0.56 |
ENSDART00000154570
|
vsig10l
|
V-set and immunoglobulin domain containing 10 like |
| chr11_-_30634286 | 0.56 |
ENSDART00000191019
|
zgc:153665
|
zgc:153665 |
| chr19_-_47526737 | 0.55 |
ENSDART00000186636
|
scg5
|
secretogranin V |
| chr16_-_41131578 | 0.55 |
ENSDART00000102649
ENSDART00000145956 |
ptpn23a
|
protein tyrosine phosphatase, non-receptor type 23, a |
| chr1_+_49668423 | 0.54 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr5_-_28029558 | 0.54 |
ENSDART00000078649
|
abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr5_+_26913120 | 0.54 |
ENSDART00000126609
|
tbx3b
|
T-box 3b |
| chr6_+_3934738 | 0.53 |
ENSDART00000159673
|
dync1i2b
|
dynein, cytoplasmic 1, intermediate chain 2b |
| chr15_-_34558579 | 0.53 |
ENSDART00000140882
|
ispd
|
isoprenoid synthase domain containing |
| chr8_+_17184602 | 0.52 |
ENSDART00000050228
ENSDART00000140531 |
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr2_+_5563077 | 0.52 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr19_-_28789404 | 0.51 |
ENSDART00000191453
ENSDART00000026992 |
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr22_-_24285432 | 0.51 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr3_+_29941777 | 0.51 |
ENSDART00000113889
|
ifi35
|
interferon-induced protein 35 |
| chr11_+_2416064 | 0.49 |
ENSDART00000067117
|
ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr12_+_1455147 | 0.49 |
ENSDART00000018752
|
cops3
|
COP9 signalosome subunit 3 |
| chr2_+_10134345 | 0.49 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr25_+_14087045 | 0.48 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr1_+_2190714 | 0.48 |
ENSDART00000132126
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr3_+_52442102 | 0.47 |
ENSDART00000155455
|
si:ch211-191c10.1
|
si:ch211-191c10.1 |
| chr21_+_30549512 | 0.47 |
ENSDART00000132831
|
rab38c
|
RAB38c, member of RAS oncogene family |
| chr7_-_27033080 | 0.47 |
ENSDART00000173516
|
nucb2a
|
nucleobindin 2a |
| chr23_+_36653376 | 0.45 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr13_-_47403154 | 0.44 |
ENSDART00000114318
|
bcl2l11
|
BCL2 like 11 |
| chr2_+_16160906 | 0.44 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr8_-_14184423 | 0.43 |
ENSDART00000063817
|
ndufb11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 |
| chr3_-_41292275 | 0.42 |
ENSDART00000144088
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr4_+_1600034 | 0.42 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr16_-_34477805 | 0.42 |
ENSDART00000136546
|
serinc2l
|
serine incorporator 2, like |
| chr22_+_31821815 | 0.41 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr4_-_6373735 | 0.41 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr17_+_6079881 | 0.41 |
ENSDART00000033947
|
chrna2a
|
cholinergic receptor, nicotinic, alpha 2a (neuronal) |
| chr1_+_25801648 | 0.41 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr5_+_34622320 | 0.41 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr24_+_25259154 | 0.40 |
ENSDART00000171125
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr24_-_26369185 | 0.40 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr11_-_12705608 | 0.40 |
ENSDART00000158937
ENSDART00000114391 |
zgc:174353
|
zgc:174353 |
| chr19_-_47527093 | 0.40 |
ENSDART00000171379
ENSDART00000171140 ENSDART00000114886 |
scg5
|
secretogranin V |
| chr5_+_36513605 | 0.39 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr3_+_13559199 | 0.39 |
ENSDART00000166547
|
si:ch73-106n3.1
|
si:ch73-106n3.1 |
| chr20_+_51104367 | 0.38 |
ENSDART00000073981
|
eif2s1b
|
eukaryotic translation initiation factor 2, subunit 1 alpha b |
| chr24_-_31306724 | 0.38 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr23_-_44848961 | 0.38 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr3_+_32492467 | 0.38 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr20_+_6142433 | 0.38 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr9_-_456541 | 0.37 |
ENSDART00000159596
|
zgc:91849
|
zgc:91849 |
| chr1_-_44940830 | 0.37 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr13_-_36439061 | 0.36 |
ENSDART00000084867
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr18_-_16795262 | 0.36 |
ENSDART00000048722
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr8_+_26818446 | 0.35 |
ENSDART00000134987
ENSDART00000138835 |
si:ch211-156j16.1
|
si:ch211-156j16.1 |
| chr4_-_18436899 | 0.35 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr4_-_64703 | 0.35 |
ENSDART00000167851
|
CU856344.1
|
|
| chr7_-_26844064 | 0.34 |
ENSDART00000162241
|
si:ch211-107p11.3
|
si:ch211-107p11.3 |
| chr3_+_5575313 | 0.34 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr17_+_51262556 | 0.34 |
ENSDART00000186748
ENSDART00000181606 ENSDART00000063738 ENSDART00000189066 |
eipr1
|
EARP complex and GARP complex interacting protein 1 |
| chr4_-_14926637 | 0.34 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr21_+_17768174 | 0.33 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr21_-_20840714 | 0.32 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr22_+_6905210 | 0.32 |
ENSDART00000167530
|
CABZ01065328.2
|
|
| chr21_-_22951604 | 0.32 |
ENSDART00000083449
ENSDART00000180129 |
dub
|
duboraya |
| chr19_+_19600297 | 0.32 |
ENSDART00000160134
ENSDART00000183493 |
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr3_-_41292569 | 0.32 |
ENSDART00000111856
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr25_-_31423493 | 0.32 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr18_+_17611627 | 0.31 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr5_-_66160415 | 0.31 |
ENSDART00000073895
|
mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr15_+_15390882 | 0.31 |
ENSDART00000062024
|
ca4b
|
carbonic anhydrase IV b |
| chr21_-_25573064 | 0.30 |
ENSDART00000134310
|
CR388166.1
|
|
| chr6_-_8736766 | 0.30 |
ENSDART00000143956
|
cavin2b
|
caveolae associated protein 2b |
| chr24_+_23716918 | 0.29 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr8_+_48484455 | 0.29 |
ENSDART00000122737
|
PRDM16
|
si:ch211-263k4.2 |
| chr3_-_6719232 | 0.28 |
ENSDART00000154294
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr17_+_8324345 | 0.28 |
ENSDART00000172443
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr4_-_1824836 | 0.28 |
ENSDART00000111858
|
mrpl42
|
mitochondrial ribosomal protein L42 |
| chr22_-_21176269 | 0.27 |
ENSDART00000112839
|
rex1bd
|
required for excision 1-B domain containing |
| chr10_+_19017146 | 0.27 |
ENSDART00000038674
|
tmem230a
|
transmembrane protein 230a |
| chr17_-_53439866 | 0.27 |
ENSDART00000154826
|
mycbp
|
c-myc binding protein |
| chr15_-_28247583 | 0.26 |
ENSDART00000112967
|
rilp
|
Rab interacting lysosomal protein |
| chr16_+_11724230 | 0.26 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr8_+_8012570 | 0.26 |
ENSDART00000183429
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr24_-_26485098 | 0.26 |
ENSDART00000135496
ENSDART00000009609 ENSDART00000133782 ENSDART00000141029 ENSDART00000113739 |
eif5a
|
eukaryotic translation initiation factor 5A |
| chr12_-_32066469 | 0.26 |
ENSDART00000140685
ENSDART00000062185 |
rab40b
|
RAB40B, member RAS oncogene family |
| chr16_+_43152727 | 0.26 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr10_-_25069155 | 0.25 |
ENSDART00000078226
ENSDART00000181941 |
mtnr1bb
|
melatonin receptor 1Bb |
| chr11_-_36350876 | 0.25 |
ENSDART00000146495
ENSDART00000020655 |
psma5
|
proteasome subunit alpha 5 |
| chr3_+_12484008 | 0.25 |
ENSDART00000182229
|
vasnb
|
vasorin b |
| chr20_+_38525567 | 0.25 |
ENSDART00000147787
|
znf512
|
zinc finger protein 512 |
| chr15_+_29123031 | 0.25 |
ENSDART00000133988
ENSDART00000060030 |
zgc:101731
|
zgc:101731 |
| chr8_+_39795918 | 0.25 |
ENSDART00000143413
|
si:ch211-170d8.2
|
si:ch211-170d8.2 |
| chr11_-_36350421 | 0.25 |
ENSDART00000141477
|
psma5
|
proteasome subunit alpha 5 |
| chr2_+_38161318 | 0.25 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr20_+_46492175 | 0.24 |
ENSDART00000060695
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr11_+_7528599 | 0.24 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr21_-_39081107 | 0.24 |
ENSDART00000075935
|
vtnb
|
vitronectin b |
| chr7_-_18545515 | 0.23 |
ENSDART00000040534
|
rgs12a
|
regulator of G protein signaling 12a |
| chr24_+_11381400 | 0.23 |
ENSDART00000058703
|
ackr4b
|
atypical chemokine receptor 4b |
| chr19_-_6134802 | 0.23 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr9_+_13714379 | 0.23 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr23_+_24600494 | 0.23 |
ENSDART00000184854
|
nckap5l
|
NCK-associated protein 5-like |
| chr2_-_11258547 | 0.23 |
ENSDART00000165803
ENSDART00000193817 |
slc44a5a
|
solute carrier family 44, member 5a |
| chr8_+_12930216 | 0.23 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr9_+_44430974 | 0.23 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr9_-_34396264 | 0.23 |
ENSDART00000045754
|
grtp1b
|
growth hormone regulated TBC protein 1b |
| chr11_-_31172276 | 0.22 |
ENSDART00000171520
|
si:dkey-238i5.3
|
si:dkey-238i5.3 |
| chr5_+_32345187 | 0.22 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr25_+_4812685 | 0.22 |
ENSDART00000193103
|
myo5c
|
myosin VC |
| chr6_+_41446541 | 0.22 |
ENSDART00000029553
ENSDART00000128756 ENSDART00000144864 |
rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr9_-_21912227 | 0.22 |
ENSDART00000145576
|
lmo7a
|
LIM domain 7a |
| chr23_+_32044410 | 0.22 |
ENSDART00000048628
|
mylk2
|
myosin light chain kinase 2 |
| chr20_+_38620681 | 0.22 |
ENSDART00000045850
|
si:ch211-243j20.2
|
si:ch211-243j20.2 |
| chr15_+_36187434 | 0.22 |
ENSDART00000181536
ENSDART00000099501 ENSDART00000154432 |
masp1
|
mannan-binding lectin serine peptidase 1 |
| chr15_+_24644016 | 0.21 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr20_+_54274431 | 0.21 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr5_-_24000211 | 0.21 |
ENSDART00000188865
|
map7d2a
|
MAP7 domain containing 2a |
| chr22_+_7497319 | 0.21 |
ENSDART00000034564
|
CELA1 (1 of many)
|
zgc:92511 |
| chr4_-_16334362 | 0.21 |
ENSDART00000101461
|
epyc
|
epiphycan |
| chr20_-_39273505 | 0.20 |
ENSDART00000153114
|
clu
|
clusterin |
| chr17_+_43843536 | 0.20 |
ENSDART00000164097
|
FO704777.1
|
|
| chr9_-_2573121 | 0.20 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr4_-_12286067 | 0.20 |
ENSDART00000022646
|
cnot4b
|
CCR4-NOT transcription complex, subunit 4b |
| chr6_+_18298444 | 0.20 |
ENSDART00000166018
|
card14
|
caspase recruitment domain family, member 14 |
| chr21_-_25774605 | 0.19 |
ENSDART00000002128
|
CLDN4 (1 of many)
|
zgc:112437 |
| chr14_+_20893065 | 0.19 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr7_+_39684652 | 0.19 |
ENSDART00000040946
|
tbc1d14
|
TBC1 domain family, member 14 |
| chr24_+_25258904 | 0.18 |
ENSDART00000155714
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr1_-_26071535 | 0.18 |
ENSDART00000185187
|
pdcd4a
|
programmed cell death 4a |
| chr1_+_33647682 | 0.18 |
ENSDART00000114384
|
znf654
|
zinc finger protein 654 |
| chr22_-_36856405 | 0.18 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr16_-_13109749 | 0.18 |
ENSDART00000142610
|
prkcg
|
protein kinase C, gamma |
| chr11_-_25539323 | 0.18 |
ENSDART00000155785
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr13_+_22480857 | 0.17 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr5_-_52243424 | 0.17 |
ENSDART00000159078
|
erap2
|
endoplasmic reticulum aminopeptidase 2 |
| chr16_+_18535618 | 0.17 |
ENSDART00000021596
|
rxrbb
|
retinoid x receptor, beta b |
| chr13_+_30169233 | 0.17 |
ENSDART00000181183
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr17_-_42988356 | 0.17 |
ENSDART00000024558
|
zgc:92137
|
zgc:92137 |
| chr20_+_28960340 | 0.17 |
ENSDART00000147917
|
gpr176
|
G protein-coupled receptor 176 |
| chr17_-_43399896 | 0.17 |
ENSDART00000156033
ENSDART00000156418 |
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr15_-_35100858 | 0.17 |
ENSDART00000099677
|
bloc1s3
|
biogenesis of lysosomal organelles complex-1, subunit 3 |
| chr8_-_16697912 | 0.17 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr23_+_2778813 | 0.17 |
ENSDART00000142621
|
top1
|
DNA topoisomerase I |
| chr20_-_39273987 | 0.17 |
ENSDART00000127173
|
clu
|
clusterin |
| chr3_+_32443395 | 0.17 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxa3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0090220 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.2 | 0.6 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.2 | 0.8 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 1.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.9 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.4 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 0.8 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.5 | GO:0045824 | negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.1 | 0.3 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.3 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.1 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.4 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 1.1 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.2 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.6 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.6 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.1 | 0.7 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.2 | GO:0016117 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 0.8 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 2.1 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.0 | 0.4 | GO:0009914 | hormone transport(GO:0009914) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.0 | 0.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.7 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.4 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.6 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.2 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 2.6 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.7 | GO:0039022 | pronephric duct development(GO:0039022) |
| 0.0 | 0.5 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.2 | GO:1902023 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.3 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.1 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.1 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.5 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.4 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.5 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.7 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.5 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 1.0 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 1.2 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.0 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.0 | GO:1903779 | regulation of heart looping(GO:1901207) regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.1 | GO:0030320 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 0.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.4 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.5 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 0.2 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.5 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.7 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.0 | 2.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.5 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.6 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.2 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 0.5 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.2 | 1.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.6 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 1.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.3 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.2 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.5 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 1.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |