Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
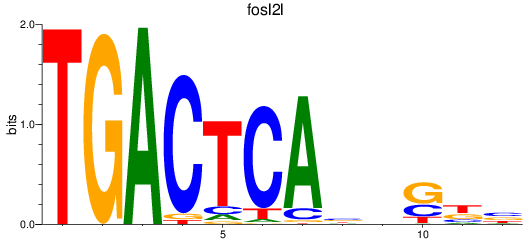
Results for fosl2l
Z-value: 0.49
Transcription factors associated with fosl2l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosl2l
|
ENSDARG00000092174 | fos-like antigen 2 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-59d15.4 | dr11_v1_chr20_+_9211237_9211237 | -0.27 | 2.8e-01 | Click! |
Activity profile of fosl2l motif
Sorted Z-values of fosl2l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_32877348 | 1.10 |
ENSDART00000018977
ENSDART00000133421 |
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr24_-_33366188 | 0.91 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr1_+_46598764 | 0.78 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr7_-_26262978 | 0.67 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr16_+_23403602 | 0.66 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr5_+_9037650 | 0.62 |
ENSDART00000158226
|
si:ch211-155m12.1
|
si:ch211-155m12.1 |
| chr1_+_46598502 | 0.62 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr8_+_8671229 | 0.61 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr19_+_37120491 | 0.59 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr19_-_7043355 | 0.57 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr6_-_33685325 | 0.57 |
ENSDART00000181883
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr7_+_66884570 | 0.56 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr22_+_17399124 | 0.51 |
ENSDART00000145769
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr12_-_3077395 | 0.49 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr5_+_34549845 | 0.48 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr20_-_34670236 | 0.48 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr2_-_985417 | 0.46 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr6_-_59357256 | 0.46 |
ENSDART00000074534
|
fam210b
|
family with sequence similarity 210, member B |
| chr20_+_23501535 | 0.45 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr9_+_33154841 | 0.44 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr24_+_24726956 | 0.44 |
ENSDART00000144574
ENSDART00000066628 |
mtfr1
|
mitochondrial fission regulator 1 |
| chr22_+_24603930 | 0.44 |
ENSDART00000180240
ENSDART00000164256 |
CU655842.1
|
|
| chr5_-_49951106 | 0.44 |
ENSDART00000135954
|
fam172a
|
family with sequence similarity 172, member A |
| chr21_+_13245302 | 0.44 |
ENSDART00000189498
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr9_+_22929675 | 0.44 |
ENSDART00000061299
|
tsn
|
translin |
| chr19_-_7070691 | 0.43 |
ENSDART00000168755
|
tapbp.2
|
TAP binding protein (tapasin), tandem duplicate 2 |
| chr5_-_13076779 | 0.42 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr23_-_24226533 | 0.42 |
ENSDART00000109134
|
plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr18_+_15841449 | 0.41 |
ENSDART00000141800
ENSDART00000091349 |
eea1
|
early endosome antigen 1 |
| chr7_-_7764287 | 0.41 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr17_+_51743908 | 0.41 |
ENSDART00000149039
ENSDART00000148869 |
odc1
|
ornithine decarboxylase 1 |
| chr12_-_17147473 | 0.40 |
ENSDART00000106035
|
stambpl1
|
STAM binding protein-like 1 |
| chr9_+_33158191 | 0.39 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr20_+_28364742 | 0.39 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr15_+_17343319 | 0.39 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr22_+_23359369 | 0.39 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr21_+_43199237 | 0.39 |
ENSDART00000151748
|
aff4
|
AF4/FMR2 family, member 4 |
| chr21_-_20945658 | 0.38 |
ENSDART00000079701
|
rnf180
|
ring finger protein 180 |
| chr16_+_13818743 | 0.38 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr4_+_20051478 | 0.38 |
ENSDART00000143642
|
lamtor4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr4_-_1801519 | 0.38 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr23_+_28378543 | 0.38 |
ENSDART00000145327
|
zgc:153867
|
zgc:153867 |
| chr5_+_51227147 | 0.37 |
ENSDART00000083340
|
ubac1
|
UBA domain containing 1 |
| chr17_+_51744450 | 0.37 |
ENSDART00000190955
ENSDART00000149807 |
odc1
|
ornithine decarboxylase 1 |
| chr4_+_25181572 | 0.36 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr12_-_5455936 | 0.35 |
ENSDART00000109305
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr16_+_13818500 | 0.35 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr19_-_5805923 | 0.34 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr12_-_35787801 | 0.33 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr9_+_28140089 | 0.33 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr2_+_37838259 | 0.33 |
ENSDART00000136796
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr22_+_26600834 | 0.32 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr10_+_16225117 | 0.32 |
ENSDART00000169885
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr25_+_3549401 | 0.32 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr4_+_5317483 | 0.31 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr6_+_51713076 | 0.30 |
ENSDART00000146281
|
ripor3
|
RIPOR family member 3 |
| chr17_+_32623931 | 0.30 |
ENSDART00000144217
|
ctsba
|
cathepsin Ba |
| chr7_+_50395856 | 0.30 |
ENSDART00000032324
|
hddc3
|
HD domain containing 3 |
| chr18_-_37355666 | 0.30 |
ENSDART00000098914
|
yap1
|
Yes-associated protein 1 |
| chr2_+_25378457 | 0.29 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr6_+_49901465 | 0.29 |
ENSDART00000023515
|
chmp4ba
|
charged multivesicular body protein 4Ba |
| chr11_-_18601955 | 0.29 |
ENSDART00000180565
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr3_+_19245804 | 0.29 |
ENSDART00000134514
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr9_+_41024973 | 0.29 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr7_-_7398350 | 0.28 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr6_+_54711306 | 0.28 |
ENSDART00000074605
|
pkp1b
|
plakophilin 1b |
| chr19_-_43750389 | 0.27 |
ENSDART00000147328
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr3_+_19687217 | 0.27 |
ENSDART00000141937
|
tlk2
|
tousled-like kinase 2 |
| chr3_-_39190317 | 0.27 |
ENSDART00000013167
|
retsat
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr4_+_1722899 | 0.27 |
ENSDART00000021139
|
fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr15_+_24704798 | 0.27 |
ENSDART00000192470
|
LRRC75A
|
si:dkey-151p21.7 |
| chr3_-_32362872 | 0.26 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr3_-_21062706 | 0.26 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr7_+_34549198 | 0.25 |
ENSDART00000173784
|
fhod1
|
formin homology 2 domain containing 1 |
| chr11_-_11965033 | 0.25 |
ENSDART00000193683
|
abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr12_-_27212596 | 0.25 |
ENSDART00000153101
|
psme3
|
proteasome activator subunit 3 |
| chr5_-_42123794 | 0.25 |
ENSDART00000051135
|
emc6
|
ER membrane protein complex subunit 6 |
| chr8_+_50190742 | 0.25 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr3_-_24205339 | 0.24 |
ENSDART00000157135
|
si:dkey-110g7.8
|
si:dkey-110g7.8 |
| chr21_+_6290566 | 0.24 |
ENSDART00000161647
|
fnbp1b
|
formin binding protein 1b |
| chr21_-_14692119 | 0.24 |
ENSDART00000123047
|
ehmt1b
|
euchromatic histone-lysine N-methyltransferase 1b |
| chr15_+_40188076 | 0.24 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr24_-_31090948 | 0.24 |
ENSDART00000176799
|
hccsb
|
holocytochrome c synthase b |
| chr15_+_20352123 | 0.23 |
ENSDART00000011030
ENSDART00000163532 ENSDART00000169537 ENSDART00000161047 |
il15l
|
interleukin 15, like |
| chr7_+_34549377 | 0.23 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr23_-_14990865 | 0.23 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr23_-_7797207 | 0.22 |
ENSDART00000181611
|
myt1b
|
myelin transcription factor 1b |
| chr25_+_18583877 | 0.22 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr19_+_48359259 | 0.22 |
ENSDART00000167353
|
sgo1
|
shugoshin 1 |
| chr23_-_36003441 | 0.22 |
ENSDART00000164699
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr1_+_41690402 | 0.21 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr13_+_6188759 | 0.21 |
ENSDART00000161062
|
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr8_-_11170114 | 0.21 |
ENSDART00000133532
|
si:ch211-204d2.4
|
si:ch211-204d2.4 |
| chr11_+_45153104 | 0.21 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr18_+_618005 | 0.21 |
ENSDART00000189667
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr15_+_2559875 | 0.21 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr8_-_18516385 | 0.21 |
ENSDART00000149446
|
si:dkey-30h22.11
|
si:dkey-30h22.11 |
| chr25_+_13498188 | 0.20 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr21_+_6328801 | 0.20 |
ENSDART00000163577
|
fnbp1b
|
formin binding protein 1b |
| chr8_+_54274355 | 0.19 |
ENSDART00000067639
|
TMCC1 (1 of many)
|
transmembrane and coiled-coil domain family 1 |
| chr23_-_7799184 | 0.19 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr12_+_32292564 | 0.19 |
ENSDART00000152945
|
ANKFN1
|
si:ch211-277e21.2 |
| chr18_-_41160975 | 0.19 |
ENSDART00000187517
|
CABZ01005876.1
|
|
| chr25_+_8407892 | 0.19 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr23_-_36003282 | 0.19 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr23_+_19759128 | 0.19 |
ENSDART00000135820
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr25_-_31907590 | 0.18 |
ENSDART00000149707
ENSDART00000149471 |
eif3ja
|
eukaryotic translation initiation factor 3, subunit Ja |
| chr5_-_20678300 | 0.18 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr24_+_19415124 | 0.18 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr17_+_43946269 | 0.18 |
ENSDART00000017376
|
ktn1
|
kinectin 1 |
| chr23_-_437467 | 0.18 |
ENSDART00000192106
|
tspan2b
|
tetraspanin 2b |
| chr15_+_29709382 | 0.18 |
ENSDART00000099956
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr13_+_6189203 | 0.17 |
ENSDART00000109665
|
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr4_-_4535189 | 0.17 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr14_-_22100118 | 0.17 |
ENSDART00000157547
|
ssrp1a
|
structure specific recognition protein 1a |
| chr3_+_24708685 | 0.17 |
ENSDART00000153507
|
mkl1a
|
megakaryoblastic leukemia (translocation) 1a |
| chr25_+_10410620 | 0.17 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr7_-_40145097 | 0.17 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr19_+_49721 | 0.17 |
ENSDART00000160489
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr5_-_23464970 | 0.17 |
ENSDART00000141028
|
si:dkeyp-20g2.1
|
si:dkeyp-20g2.1 |
| chr25_-_6432463 | 0.17 |
ENSDART00000110389
|
ptpn9a
|
protein tyrosine phosphatase, non-receptor type 9, a |
| chr4_-_789645 | 0.17 |
ENSDART00000164441
|
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr17_+_24597001 | 0.16 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr5_+_24059598 | 0.16 |
ENSDART00000051547
|
gabarapa
|
GABA(A) receptor-associated protein a |
| chr19_+_15441022 | 0.16 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr6_-_60079551 | 0.16 |
ENSDART00000154753
|
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr3_-_30248277 | 0.16 |
ENSDART00000133516
ENSDART00000077029 |
emc10
|
ER membrane protein complex subunit 10 |
| chr7_+_67381912 | 0.16 |
ENSDART00000167564
|
nfat5b
|
nuclear factor of activated T cells 5b |
| chr9_-_12493628 | 0.16 |
ENSDART00000180832
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr10_+_11260170 | 0.16 |
ENSDART00000155742
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr23_-_1017428 | 0.16 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr9_+_46745348 | 0.15 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr23_-_31693309 | 0.15 |
ENSDART00000146327
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr6_-_25189739 | 0.15 |
ENSDART00000181980
|
lrrc8db
|
leucine rich repeat containing 8 VRAC subunit Db |
| chr13_-_33134611 | 0.15 |
ENSDART00000026280
|
plek2
|
pleckstrin 2 |
| chr24_+_80653 | 0.15 |
ENSDART00000158473
ENSDART00000129135 |
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr5_+_19320554 | 0.15 |
ENSDART00000165119
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr13_+_19283936 | 0.15 |
ENSDART00000111462
|
eif3s10
|
eukaryotic translation initiation factor 3, subunit 10 (theta) |
| chr10_-_105100 | 0.15 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr25_+_32496723 | 0.15 |
ENSDART00000087978
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr19_-_43757568 | 0.14 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr7_-_40144686 | 0.14 |
ENSDART00000109166
|
wdr60
|
WD repeat domain 60 |
| chr6_+_18367388 | 0.14 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr4_+_9400012 | 0.14 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr21_-_30254185 | 0.14 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr8_-_22288004 | 0.14 |
ENSDART00000100042
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr12_+_1286642 | 0.14 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr23_+_24989387 | 0.14 |
ENSDART00000172299
ENSDART00000145307 |
arhgap4a
|
Rho GTPase activating protein 4a |
| chr7_+_7019911 | 0.14 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr19_-_7420867 | 0.14 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr8_-_22288258 | 0.13 |
ENSDART00000140978
ENSDART00000100046 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr10_-_13239367 | 0.13 |
ENSDART00000001253
|
si:busm1-57f23.1
|
si:busm1-57f23.1 |
| chr5_+_61556172 | 0.13 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr23_+_33718602 | 0.13 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr21_+_17301790 | 0.13 |
ENSDART00000145057
|
tsc1b
|
TSC complex subunit 1b |
| chr8_+_10835456 | 0.12 |
ENSDART00000151388
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr23_+_30967686 | 0.12 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr4_-_4261673 | 0.12 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr20_-_21994901 | 0.12 |
ENSDART00000004984
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr7_-_44704910 | 0.12 |
ENSDART00000037850
|
dync1li2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr11_+_11267829 | 0.12 |
ENSDART00000026814
ENSDART00000173346 ENSDART00000151926 |
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr23_-_4704938 | 0.12 |
ENSDART00000067293
|
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr22_+_5574952 | 0.11 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr2_-_39675829 | 0.11 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr2_+_47944967 | 0.11 |
ENSDART00000135584
|
ftr19
|
finTRIM family, member 19 |
| chr7_+_38680034 | 0.11 |
ENSDART00000171691
ENSDART00000007913 |
psmc3
|
proteasome 26S subunit, ATPase 3 |
| chr3_+_43955864 | 0.10 |
ENSDART00000168267
|
BX571715.1
|
|
| chr22_-_38543630 | 0.10 |
ENSDART00000172029
|
si:ch211-126j24.1
|
si:ch211-126j24.1 |
| chr7_-_71456117 | 0.10 |
ENSDART00000042492
|
sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr22_+_17828267 | 0.10 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr11_-_27821 | 0.10 |
ENSDART00000158769
ENSDART00000172970 ENSDART00000173118 ENSDART00000168674 ENSDART00000163545 ENSDART00000173411 ENSDART00000172132 |
sp1
|
sp1 transcription factor |
| chr15_-_43978141 | 0.10 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr9_+_38481780 | 0.09 |
ENSDART00000087241
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr14_+_23184517 | 0.09 |
ENSDART00000181410
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr23_-_1017605 | 0.09 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr2_+_47945359 | 0.09 |
ENSDART00000098054
|
ftr19
|
finTRIM family, member 19 |
| chr14_-_16807206 | 0.09 |
ENSDART00000157957
|
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr23_+_25292147 | 0.08 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr20_-_47347962 | 0.08 |
ENSDART00000080863
|
dtnba
|
dystrobrevin, beta a |
| chr18_-_13360106 | 0.08 |
ENSDART00000091512
|
cmip
|
c-Maf inducing protein |
| chr20_-_47348116 | 0.08 |
ENSDART00000162087
ENSDART00000160769 ENSDART00000164484 |
dtnba
|
dystrobrevin, beta a |
| chr11_-_14102131 | 0.08 |
ENSDART00000085158
ENSDART00000191962 |
tmem259
|
transmembrane protein 259 |
| chr9_-_9282519 | 0.08 |
ENSDART00000146821
|
si:ch211-214p13.3
|
si:ch211-214p13.3 |
| chr11_+_11267493 | 0.08 |
ENSDART00000148425
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr20_+_37661229 | 0.07 |
ENSDART00000138539
|
aig1
|
androgen-induced 1 (H. sapiens) |
| chr2_+_27932781 | 0.07 |
ENSDART00000019555
|
mc4r
|
melanocortin 4 receptor |
| chr23_-_4705110 | 0.07 |
ENSDART00000144536
ENSDART00000129050 ENSDART00000136399 |
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr2_-_24407933 | 0.07 |
ENSDART00000088584
|
si:dkey-208k22.6
|
si:dkey-208k22.6 |
| chr17_-_45150763 | 0.07 |
ENSDART00000155043
ENSDART00000156786 ENSDART00000191147 |
tmed8
|
transmembrane p24 trafficking protein 8 |
| chr15_+_42235449 | 0.07 |
ENSDART00000114801
ENSDART00000182053 |
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr11_-_45152702 | 0.07 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr17_+_28624321 | 0.07 |
ENSDART00000122260
|
heatr5a
|
HEAT repeat containing 5a |
| chr6_-_41091151 | 0.07 |
ENSDART00000154963
ENSDART00000153818 |
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr10_-_44482911 | 0.07 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr9_-_47654204 | 0.07 |
ENSDART00000015159
|
tns1b
|
tensin 1b |
| chr16_-_31598771 | 0.06 |
ENSDART00000016386
|
tg
|
thyroglobulin |
| chr8_+_29749017 | 0.06 |
ENSDART00000185144
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr9_+_38962398 | 0.06 |
ENSDART00000134294
|
map2
|
microtubule-associated protein 2 |
| chr16_+_48714048 | 0.06 |
ENSDART00000148709
ENSDART00000150121 |
brd2b
|
bromodomain containing 2b |
| chr4_+_77661056 | 0.06 |
ENSDART00000152953
|
si:dkey-61p9.7
|
si:dkey-61p9.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of fosl2l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.1 | 0.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.6 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.7 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.4 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 0.8 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.4 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.3 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.2 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.6 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.3 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.4 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.3 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.1 | 0.4 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 0.2 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 0.6 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.5 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.3 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.3 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.4 | GO:1903729 | regulation of plasma membrane organization(GO:1903729) |
| 0.0 | 0.4 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.5 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.1 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.3 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.2 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.1 | GO:0048521 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.0 | GO:1901296 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.4 | GO:0007164 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.4 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.6 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.8 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 0.4 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.4 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 1.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.3 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |