Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
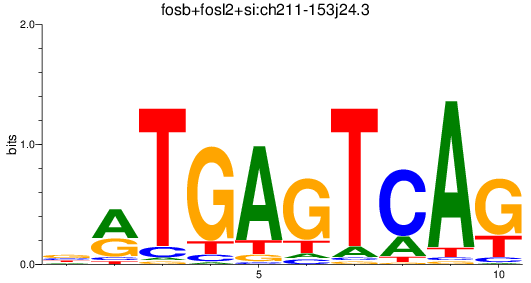
Results for fosb+fosl2+si:ch211-153j24.3
Z-value: 0.85
Transcription factors associated with fosb+fosl2+si:ch211-153j24.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosl2
|
ENSDARG00000040623 | fos-like antigen 2 |
|
fosb
|
ENSDARG00000055751 | FBJ murine osteosarcoma viral oncogene homolog B |
|
si_ch211-153j24.3
|
ENSDARG00000068428 | si_ch211-153j24.3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| fosb | dr11_v1_chr18_+_36770166_36770166 | 0.91 | 1.5e-07 | Click! |
| fosl2 | dr11_v1_chr17_+_41302660_41302660 | 0.81 | 4.3e-05 | Click! |
| si:ch211-153j24.3 | dr11_v1_chr20_+_46560258_46560258 | 0.52 | 2.5e-02 | Click! |
Activity profile of fosb+fosl2+si:ch211-153j24.3 motif
Sorted Z-values of fosb+fosl2+si:ch211-153j24.3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_46382484 | 3.49 |
ENSDART00000024202
ENSDART00000142790 |
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr12_-_212843 | 3.47 |
ENSDART00000083574
|
CABZ01102039.1
|
|
| chr11_-_24681292 | 3.30 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr10_+_31222656 | 2.48 |
ENSDART00000140988
ENSDART00000143387 |
tmem218
|
transmembrane protein 218 |
| chr16_+_42829735 | 2.39 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr24_+_31361407 | 2.26 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr10_+_31222433 | 1.93 |
ENSDART00000185080
|
tmem218
|
transmembrane protein 218 |
| chr2_+_17524278 | 1.85 |
ENSDART00000165633
|
pimr196
|
Pim proto-oncogene, serine/threonine kinase, related 196 |
| chr5_-_21888368 | 1.83 |
ENSDART00000020725
|
smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr3_-_61593274 | 1.73 |
ENSDART00000154132
ENSDART00000055071 |
nptx2a
|
neuronal pentraxin 2a |
| chr21_-_30545121 | 1.65 |
ENSDART00000019199
|
rab39ba
|
RAB39B, member RAS oncogene family a |
| chr1_-_52498146 | 1.64 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr7_-_52963493 | 1.53 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr21_+_22828500 | 1.52 |
ENSDART00000151109
|
si:rp71-1p14.7
|
si:rp71-1p14.7 |
| chr9_-_27805801 | 1.51 |
ENSDART00000140608
ENSDART00000114542 |
si:rp71-45g20.10
|
si:rp71-45g20.10 |
| chr12_+_8074343 | 1.50 |
ENSDART00000124084
|
cabcoco1
|
ciliary associated calcium binding coiled-coil 1 |
| chr23_-_39636195 | 1.49 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr1_+_44360973 | 1.40 |
ENSDART00000167568
|
si:ch211-165a10.5
|
si:ch211-165a10.5 |
| chr16_+_42830152 | 1.40 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr10_+_26747755 | 1.30 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr10_-_27566481 | 1.27 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr16_-_13818061 | 1.26 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr6_+_29791164 | 1.25 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr23_-_37113396 | 1.25 |
ENSDART00000102886
ENSDART00000134461 |
zgc:193690
|
zgc:193690 |
| chr3_+_24986145 | 1.24 |
ENSDART00000055428
|
cbx7a
|
chromobox homolog 7a |
| chr16_+_41517188 | 1.23 |
ENSDART00000049976
|
si:dkey-11p23.7
|
si:dkey-11p23.7 |
| chr16_+_23960744 | 1.23 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr10_+_4987766 | 1.17 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr22_+_25049563 | 1.16 |
ENSDART00000078173
|
dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr1_-_59411901 | 1.14 |
ENSDART00000167015
|
si:ch211-188p14.4
|
si:ch211-188p14.4 |
| chr12_+_20506197 | 1.14 |
ENSDART00000153010
|
si:zfos-754c12.2
|
si:zfos-754c12.2 |
| chr17_+_7524389 | 1.14 |
ENSDART00000067579
|
klhl10b.2
|
kelch-like family member 10b, tandem duplicate 2 |
| chr3_+_24197934 | 1.13 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr1_-_52497834 | 1.13 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr7_-_25697285 | 1.13 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr25_+_4954220 | 1.11 |
ENSDART00000156034
|
si:ch73-265h17.2
|
si:ch73-265h17.2 |
| chr2_+_48288461 | 1.10 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr24_+_38522254 | 1.07 |
ENSDART00000156189
|
si:ch1073-66l23.1
|
si:ch1073-66l23.1 |
| chr23_-_37113215 | 1.06 |
ENSDART00000146835
|
zgc:193690
|
zgc:193690 |
| chr13_+_50368668 | 1.04 |
ENSDART00000127610
|
dnajc12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr1_+_44249616 | 0.99 |
ENSDART00000164173
|
si:ch211-165a10.10
|
si:ch211-165a10.10 |
| chr2_-_17828279 | 0.98 |
ENSDART00000083314
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr17_+_6667441 | 0.97 |
ENSDART00000123503
ENSDART00000180381 ENSDART00000156140 |
smc6
|
structural maintenance of chromosomes 6 |
| chr2_-_17827983 | 0.97 |
ENSDART00000166518
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr4_-_14915268 | 0.94 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr1_+_2190714 | 0.94 |
ENSDART00000132126
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr3_+_26064091 | 0.92 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr2_-_39759059 | 0.89 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr19_+_23932259 | 0.89 |
ENSDART00000139040
|
si:dkey-222b8.1
|
si:dkey-222b8.1 |
| chr2_+_17540720 | 0.87 |
ENSDART00000114638
|
pimr198
|
Pim proto-oncogene, serine/threonine kinase, related 198 |
| chr22_-_10110959 | 0.87 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr23_-_42752387 | 0.87 |
ENSDART00000149781
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr9_-_27771339 | 0.83 |
ENSDART00000135722
ENSDART00000140381 |
si:rp71-45g20.11
|
si:rp71-45g20.11 |
| chr22_+_16497670 | 0.83 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr3_-_8765165 | 0.80 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr2_-_14798295 | 0.78 |
ENSDART00000143430
ENSDART00000145869 |
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr11_+_13630107 | 0.76 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr25_+_21832938 | 0.74 |
ENSDART00000148299
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr23_+_3731375 | 0.74 |
ENSDART00000141782
|
smim29
|
small integral membrane protein 29 |
| chr16_-_21785261 | 0.74 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr22_-_15593824 | 0.72 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr21_+_40589770 | 0.72 |
ENSDART00000164650
ENSDART00000161584 ENSDART00000161108 |
pdk3b
|
pyruvate dehydrogenase kinase, isozyme 3b |
| chr14_+_33882973 | 0.72 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr1_-_47071979 | 0.71 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr4_-_69189894 | 0.71 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr14_-_26436760 | 0.70 |
ENSDART00000088677
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr14_-_26436951 | 0.70 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr6_+_33885828 | 0.70 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr18_+_14477740 | 0.70 |
ENSDART00000146472
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr5_-_19052184 | 0.70 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr20_-_26532167 | 0.69 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr23_+_35714574 | 0.69 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr1_+_52518176 | 0.68 |
ENSDART00000003278
|
tacr3l
|
tachykinin receptor 3-like |
| chr1_-_59139599 | 0.68 |
ENSDART00000152233
|
si:ch1073-110a20.3
|
si:ch1073-110a20.3 |
| chr4_+_357810 | 0.68 |
ENSDART00000163436
ENSDART00000103645 |
tmem181
|
transmembrane protein 181 |
| chr6_-_40899618 | 0.67 |
ENSDART00000153949
ENSDART00000021969 |
zgc:172271
|
zgc:172271 |
| chr9_-_34915351 | 0.67 |
ENSDART00000100728
ENSDART00000139608 |
upf3a
|
UPF3A, regulator of nonsense mediated mRNA decay |
| chr4_-_2440175 | 0.67 |
ENSDART00000058059
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr16_+_49005321 | 0.64 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr5_-_25582721 | 0.63 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr25_+_22643954 | 0.63 |
ENSDART00000182843
ENSDART00000121791 |
ush1c
|
Usher syndrome 1C |
| chr14_+_8725216 | 0.63 |
ENSDART00000157630
|
pimr57
|
Pim proto-oncogene, serine/threonine kinase, related 57 |
| chr4_+_16715267 | 0.62 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr23_+_4324625 | 0.62 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 ENSDART00000179819 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr9_-_27805644 | 0.61 |
ENSDART00000192431
|
si:rp71-45g20.10
|
si:rp71-45g20.10 |
| chr2_-_49997055 | 0.60 |
ENSDART00000140294
|
si:ch211-106n13.3
|
si:ch211-106n13.3 |
| chr8_+_44714336 | 0.60 |
ENSDART00000145801
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr5_+_15819651 | 0.60 |
ENSDART00000081230
ENSDART00000186969 ENSDART00000134206 |
hspb8
|
heat shock protein b8 |
| chr25_+_7423770 | 0.60 |
ENSDART00000155458
|
ubap1la
|
ubiquitin associated protein 1-like a |
| chr7_-_29200090 | 0.60 |
ENSDART00000173973
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr20_-_26531850 | 0.58 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr11_+_26609110 | 0.58 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr5_+_54400971 | 0.56 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr14_+_49382180 | 0.55 |
ENSDART00000158329
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr19_-_12078583 | 0.53 |
ENSDART00000024193
|
rnf19a
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr19_-_31402429 | 0.52 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr3_+_25825043 | 0.52 |
ENSDART00000153749
|
pik3r6b
|
phosphoinositide-3-kinase, regulatory subunit 6b |
| chr8_+_3530761 | 0.51 |
ENSDART00000081272
|
gcn1
|
GCN1 eIF2 alpha kinase activator homolog |
| chr2_+_42236118 | 0.51 |
ENSDART00000114162
ENSDART00000141199 |
ftr02
|
finTRIM family, member 2 |
| chr15_-_30832171 | 0.50 |
ENSDART00000152414
ENSDART00000152790 |
msi2b
|
musashi RNA-binding protein 2b |
| chr18_-_18849916 | 0.50 |
ENSDART00000142422
|
tgm2l
|
transglutaminase 2, like |
| chr21_+_22845317 | 0.50 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr6_-_442163 | 0.49 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr18_+_50089000 | 0.49 |
ENSDART00000058936
|
scamp5b
|
secretory carrier membrane protein 5b |
| chr24_+_39186940 | 0.48 |
ENSDART00000155817
|
spsb3b
|
splA/ryanodine receptor domain and SOCS box containing 3b |
| chr16_-_21668082 | 0.46 |
ENSDART00000088513
|
gnl1
|
guanine nucleotide binding protein-like 1 |
| chr23_+_33718602 | 0.46 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr1_-_33380340 | 0.46 |
ENSDART00000181515
|
cd99
|
CD99 molecule |
| chr5_+_38276582 | 0.45 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr25_-_3549321 | 0.45 |
ENSDART00000181214
ENSDART00000160600 |
hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr7_+_23577723 | 0.45 |
ENSDART00000049885
|
si:dkey-172j4.3
|
si:dkey-172j4.3 |
| chr25_-_173165 | 0.44 |
ENSDART00000193594
|
CABZ01114053.1
|
|
| chr21_+_30502002 | 0.44 |
ENSDART00000043727
|
slc7a3a
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3a |
| chr20_-_54508650 | 0.43 |
ENSDART00000147642
|
CABZ01118770.1
|
|
| chr19_+_37118547 | 0.42 |
ENSDART00000103163
|
cx30.9
|
connexin 30.9 |
| chr16_+_9762261 | 0.42 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr5_+_21225017 | 0.42 |
ENSDART00000141573
|
si:dkey-13n15.11
|
si:dkey-13n15.11 |
| chr4_-_13902188 | 0.42 |
ENSDART00000032805
|
gxylt1b
|
glucoside xylosyltransferase 1b |
| chr2_-_41861040 | 0.41 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr12_-_28363111 | 0.41 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr21_+_5636008 | 0.41 |
ENSDART00000158385
|
shroom3
|
shroom family member 3 |
| chr21_-_2238277 | 0.40 |
ENSDART00000165754
|
si:dkey-50i6.6
|
si:dkey-50i6.6 |
| chr14_+_34966598 | 0.40 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr2_+_7849890 | 0.39 |
ENSDART00000114241
|
si:ch211-38m6.6
|
si:ch211-38m6.6 |
| chr22_+_36547670 | 0.39 |
ENSDART00000125991
ENSDART00000186372 |
armc9
|
armadillo repeat containing 9 |
| chr9_-_710896 | 0.39 |
ENSDART00000180478
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr20_+_7084154 | 0.39 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr14_-_33613794 | 0.38 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr19_+_22216778 | 0.38 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr21_+_10794914 | 0.38 |
ENSDART00000084035
|
znf532
|
zinc finger protein 532 |
| chr6_-_41091151 | 0.38 |
ENSDART00000154963
ENSDART00000153818 |
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr24_-_6078222 | 0.37 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr1_+_41588170 | 0.37 |
ENSDART00000139175
|
si:dkey-56e3.2
|
si:dkey-56e3.2 |
| chr11_+_11267829 | 0.37 |
ENSDART00000026814
ENSDART00000173346 ENSDART00000151926 |
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr12_-_36521947 | 0.37 |
ENSDART00000152946
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr3_+_19207176 | 0.37 |
ENSDART00000087803
|
rln3a
|
relaxin 3a |
| chr2_+_21486529 | 0.36 |
ENSDART00000047468
|
inhbab
|
inhibin, beta Ab |
| chr13_-_11035420 | 0.36 |
ENSDART00000108709
|
cep170aa
|
centrosomal protein 170Aa |
| chr7_+_23875269 | 0.36 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr6_+_13933464 | 0.35 |
ENSDART00000109144
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr23_+_40460333 | 0.35 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr4_-_25168458 | 0.34 |
ENSDART00000066929
|
taf3
|
TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated facto |
| chr8_+_50983551 | 0.34 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr19_-_7495006 | 0.33 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr2_-_985417 | 0.33 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr6_-_25189739 | 0.33 |
ENSDART00000181980
|
lrrc8db
|
leucine rich repeat containing 8 VRAC subunit Db |
| chr15_-_37733238 | 0.33 |
ENSDART00000115178
|
si:dkey-42l23.7
|
si:dkey-42l23.7 |
| chr16_-_28878080 | 0.33 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr4_+_47656992 | 0.32 |
ENSDART00000161148
|
znf1040
|
zinc finger protein 1040 |
| chr10_+_28428222 | 0.32 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr4_+_5317483 | 0.32 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr13_+_6086730 | 0.31 |
ENSDART00000049328
|
fam120b
|
family with sequence similarity 120B |
| chr5_+_61556172 | 0.30 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr21_+_6328801 | 0.30 |
ENSDART00000163577
|
fnbp1b
|
formin binding protein 1b |
| chr4_-_20208166 | 0.30 |
ENSDART00000066891
|
gsap
|
gamma-secretase activating protein |
| chr5_-_43959972 | 0.29 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr7_+_13418812 | 0.29 |
ENSDART00000191905
ENSDART00000091567 |
dagla
|
diacylglycerol lipase, alpha |
| chr25_+_13191615 | 0.28 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr11_+_13629528 | 0.28 |
ENSDART00000186509
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr7_+_33172066 | 0.27 |
ENSDART00000174013
|
si:ch211-194p6.12
|
si:ch211-194p6.12 |
| chr25_+_3549401 | 0.27 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr23_+_24272421 | 0.27 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr4_+_77971104 | 0.27 |
ENSDART00000188609
|
zgc:113921
|
zgc:113921 |
| chr19_+_9050852 | 0.27 |
ENSDART00000151031
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr2_-_32366287 | 0.27 |
ENSDART00000144758
|
ubtfl
|
upstream binding transcription factor, like |
| chr2_-_22927958 | 0.26 |
ENSDART00000141621
|
myo7bb
|
myosin VIIBb |
| chr18_+_5137241 | 0.26 |
ENSDART00000159601
|
serpini1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr25_+_21324588 | 0.25 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr8_-_52871056 | 0.25 |
ENSDART00000170179
|
nr5a1a
|
nuclear receptor subfamily 5, group A, member 1a |
| chr8_-_20291922 | 0.25 |
ENSDART00000148304
|
myo1f
|
myosin IF |
| chr10_-_300000 | 0.25 |
ENSDART00000183273
|
emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr7_+_50395856 | 0.25 |
ENSDART00000032324
|
hddc3
|
HD domain containing 3 |
| chr3_-_10621391 | 0.25 |
ENSDART00000058834
|
map2k4a
|
mitogen-activated protein kinase kinase 4a |
| chr15_-_41438821 | 0.25 |
ENSDART00000136952
|
nlrc8
|
NLR family CARD domain containing 8 |
| chr3_-_32859335 | 0.24 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr10_+_1638876 | 0.24 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr17_+_15413412 | 0.24 |
ENSDART00000149871
|
cx40.8
|
connexin 40.8 |
| chr15_+_2421729 | 0.23 |
ENSDART00000082294
ENSDART00000156428 |
hephl1a
|
hephaestin-like 1a |
| chr25_+_13191391 | 0.23 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr4_-_28958601 | 0.23 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr5_-_9073433 | 0.23 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr4_+_77933084 | 0.23 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr8_+_20950401 | 0.23 |
ENSDART00000136484
ENSDART00000131329 ENSDART00000144451 ENSDART00000144929 |
si:dkeyp-82a1.1
|
si:dkeyp-82a1.1 |
| chr4_-_19742300 | 0.23 |
ENSDART00000066964
ENSDART00000100952 |
hgfa
|
hepatocyte growth factor a |
| chr3_-_61185746 | 0.22 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr9_-_21838045 | 0.22 |
ENSDART00000147471
|
acod1
|
aconitate decarboxylase 1 |
| chr25_-_11026907 | 0.22 |
ENSDART00000156846
|
mespbb
|
mesoderm posterior bb |
| chr3_-_34069637 | 0.22 |
ENSDART00000151588
|
ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr10_+_466926 | 0.22 |
ENSDART00000145220
|
arvcfa
|
ARVCF, delta catenin family member a |
| chr8_+_41038141 | 0.21 |
ENSDART00000075620
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr2_+_30379650 | 0.21 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr23_+_37185247 | 0.21 |
ENSDART00000146269
|
vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr1_+_8601935 | 0.20 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr16_+_20915319 | 0.20 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr19_+_42847306 | 0.20 |
ENSDART00000135164
|
pdcd6ip
|
programmed cell death 6 interacting protein |
| chr10_-_4316015 | 0.20 |
ENSDART00000109916
ENSDART00000180867 |
grin3a
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A |
| chr3_-_7436932 | 0.20 |
ENSDART00000163501
|
CR847850.1
|
|
| chr7_+_53541173 | 0.20 |
ENSDART00000159449
|
gramd2aa
|
GRAM domain containing 2Aa |
| chr9_+_41024973 | 0.20 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr2_-_3038904 | 0.19 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of fosb+fosl2+si:ch211-153j24.3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.3 | 0.9 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.3 | 1.1 | GO:0042117 | plasma membrane repair(GO:0001778) monocyte activation(GO:0042117) |
| 0.2 | 1.2 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.2 | 1.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.2 | 2.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.2 | 1.5 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 2.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.2 | 3.5 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 0.5 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 3.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.9 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.6 | GO:0045625 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.7 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 0.6 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 1.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.4 | GO:0089718 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.3 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 2.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 1.6 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.1 | 0.7 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.1 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.1 | 1.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.0 | GO:0030800 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.0 | 0.7 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.6 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.2 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.1 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.0 | 0.3 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.2 | GO:1903426 | regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.0 | 0.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.9 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 1.2 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.8 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.2 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.3 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 6.7 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.4 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.2 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.7 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.2 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.5 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.1 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.6 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 1.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 3.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 3.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 2.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.6 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 0.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 8.0 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 1.5 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 0.9 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.3 | 2.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 1.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 1.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.7 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.0 | GO:0008026 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 0.4 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.4 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 2.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.0 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.0 | 0.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |