Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
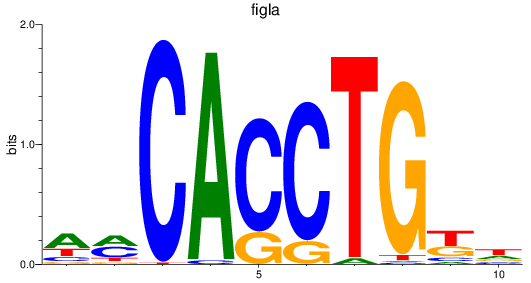
Results for figla
Z-value: 0.92
Transcription factors associated with figla
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
figla
|
ENSDARG00000087166 | folliculogenesis specific bHLH transcription factor |
|
figla
|
ENSDARG00000115572 | folliculogenesis specific bHLH transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| figla | dr11_v1_chr5_-_13848296_13848296 | 0.46 | 5.4e-02 | Click! |
Activity profile of figla motif
Sorted Z-values of figla motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_58751504 | 3.27 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr5_-_23696926 | 2.58 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr15_+_38299385 | 2.56 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr10_-_25217347 | 2.42 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr15_+_38299563 | 2.40 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr7_+_57089354 | 2.28 |
ENSDART00000140702
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr10_-_31805923 | 2.11 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr5_+_57924611 | 2.11 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr13_-_21672131 | 1.96 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr5_-_15494164 | 1.90 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr2_-_50225411 | 1.89 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr2_-_17114852 | 1.88 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr24_-_2423791 | 1.82 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr19_+_32979331 | 1.79 |
ENSDART00000078066
|
spire1a
|
spire-type actin nucleation factor 1a |
| chr7_+_57088920 | 1.68 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr2_-_17115256 | 1.67 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr7_+_67699178 | 1.65 |
ENSDART00000160086
|
zgc:162592
|
zgc:162592 |
| chr4_-_858434 | 1.61 |
ENSDART00000006961
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr14_+_24840669 | 1.46 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr6_-_37749711 | 1.42 |
ENSDART00000078324
|
nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr5_-_29531948 | 1.37 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr8_+_45334255 | 1.36 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr5_+_28271412 | 1.35 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr20_-_21672970 | 1.34 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr1_-_6085750 | 1.31 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr22_+_11756040 | 1.29 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr4_-_12978925 | 1.26 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr6_-_49537646 | 1.25 |
ENSDART00000180438
|
FO704848.1
|
|
| chr7_+_67699009 | 1.24 |
ENSDART00000192810
|
zgc:162592
|
zgc:162592 |
| chr18_+_45666489 | 1.23 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr5_-_22052852 | 1.23 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr11_-_6452444 | 1.22 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr21_-_21178410 | 1.22 |
ENSDART00000185277
ENSDART00000141341 ENSDART00000145872 ENSDART00000079678 |
ftsj1
|
FtsJ RNA methyltransferase homolog 1 |
| chr11_+_13423776 | 1.22 |
ENSDART00000102553
|
homer3b
|
homer scaffolding protein 3b |
| chr24_-_34335265 | 1.20 |
ENSDART00000128690
|
agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr13_+_39282477 | 1.19 |
ENSDART00000132198
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr3_+_26245731 | 1.18 |
ENSDART00000103734
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr10_+_37173029 | 1.17 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr14_+_35464994 | 1.17 |
ENSDART00000115307
|
si:ch211-203d1.3
|
si:ch211-203d1.3 |
| chr18_+_45573416 | 1.16 |
ENSDART00000132184
ENSDART00000145288 |
kifc3
|
kinesin family member C3 |
| chr10_-_2942900 | 1.16 |
ENSDART00000002622
|
oclna
|
occludin a |
| chr19_+_43684376 | 1.14 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr23_-_31810222 | 1.14 |
ENSDART00000134319
ENSDART00000139076 |
hbs1l
|
HBS1-like translational GTPase |
| chr22_-_17677947 | 1.14 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr23_+_9522942 | 1.12 |
ENSDART00000137751
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr18_+_45573251 | 1.09 |
ENSDART00000191309
|
kifc3
|
kinesin family member C3 |
| chr6_-_8360918 | 1.08 |
ENSDART00000004716
|
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr4_-_13614797 | 1.07 |
ENSDART00000138366
ENSDART00000165212 |
irf5
|
interferon regulatory factor 5 |
| chr2_+_1988036 | 1.07 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr5_-_65121747 | 1.05 |
ENSDART00000165556
|
tor2a
|
torsin family 2, member A |
| chr2_-_57916317 | 1.05 |
ENSDART00000183930
|
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr3_+_19665319 | 1.05 |
ENSDART00000007857
ENSDART00000193509 |
mettl2a
|
methyltransferase like 2A |
| chr20_-_13625588 | 1.04 |
ENSDART00000078893
|
sytl3
|
synaptotagmin-like 3 |
| chr14_-_31087830 | 1.03 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr16_-_20932896 | 1.03 |
ENSDART00000180646
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr21_-_4250682 | 1.02 |
ENSDART00000099389
|
dnlz
|
DNL-type zinc finger |
| chr2_+_43204919 | 1.01 |
ENSDART00000160077
ENSDART00000018729 ENSDART00000129134 ENSDART00000056402 |
pard3ab
|
par-3 family cell polarity regulator alpha, b |
| chr2_+_37838259 | 1.01 |
ENSDART00000136796
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr23_+_9522781 | 1.01 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr3_+_42923275 | 1.00 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr21_+_17016337 | 1.00 |
ENSDART00000065755
|
gpn3
|
GPN-loop GTPase 3 |
| chr19_+_7636941 | 1.00 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr1_+_594584 | 0.99 |
ENSDART00000135944
|
jam2a
|
junctional adhesion molecule 2a |
| chr3_-_26183699 | 0.99 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr12_-_23365737 | 0.98 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr1_+_20593653 | 0.97 |
ENSDART00000132440
|
si:ch211-142c4.1
|
si:ch211-142c4.1 |
| chr5_-_25572151 | 0.97 |
ENSDART00000144995
|
si:dkey-229d2.4
|
si:dkey-229d2.4 |
| chr1_+_45958904 | 0.96 |
ENSDART00000108528
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr3_-_29962345 | 0.95 |
ENSDART00000136819
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr7_-_48251234 | 0.94 |
ENSDART00000024062
ENSDART00000098904 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr19_-_10214264 | 0.93 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
| chr5_-_29152457 | 0.92 |
ENSDART00000078469
|
noxa1
|
NADPH oxidase activator 1 |
| chr14_-_35672890 | 0.91 |
ENSDART00000074710
|
pdgfc
|
platelet derived growth factor c |
| chr3_-_2613990 | 0.91 |
ENSDART00000137102
|
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr12_-_4301234 | 0.90 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr12_-_47601845 | 0.90 |
ENSDART00000169548
ENSDART00000182889 |
rgs7b
|
regulator of G protein signaling 7b |
| chr5_+_6670945 | 0.89 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr9_-_28939181 | 0.89 |
ENSDART00000101276
ENSDART00000135334 |
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr5_-_69212184 | 0.88 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr11_+_34921492 | 0.86 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr5_+_4016271 | 0.85 |
ENSDART00000113627
ENSDART00000105832 ENSDART00000121415 |
ggnbp2
|
gametogenetin binding protein 2 |
| chr1_-_53625142 | 0.84 |
ENSDART00000166852
|
USP34
|
ubiquitin specific peptidase 34 |
| chr19_-_30404096 | 0.83 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr14_+_591216 | 0.83 |
ENSDART00000169624
|
zgc:158257
|
zgc:158257 |
| chr17_+_1360192 | 0.82 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr13_+_31716820 | 0.82 |
ENSDART00000034745
|
prkcha
|
protein kinase C, eta, a |
| chr11_-_43473824 | 0.82 |
ENSDART00000179561
|
tmem63bb
|
transmembrane protein 63Bb |
| chr18_+_22220656 | 0.81 |
ENSDART00000191862
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr20_-_1191910 | 0.81 |
ENSDART00000043218
|
ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr22_-_5171829 | 0.80 |
ENSDART00000140313
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr14_-_33277743 | 0.80 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr16_+_43077909 | 0.79 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr6_-_41135215 | 0.79 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr21_+_39100289 | 0.79 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr5_+_54497730 | 0.78 |
ENSDART00000157722
|
tmem203
|
transmembrane protein 203 |
| chr14_-_33278084 | 0.78 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr2_+_27855102 | 0.78 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr5_+_54497475 | 0.78 |
ENSDART00000158149
ENSDART00000163968 ENSDART00000160446 |
tmem203
|
transmembrane protein 203 |
| chr2_+_9560740 | 0.77 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr15_+_34963316 | 0.76 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr4_-_277081 | 0.76 |
ENSDART00000166174
|
si:ch73-252i11.1
|
si:ch73-252i11.1 |
| chr15_-_29326254 | 0.76 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr11_-_44999858 | 0.76 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr6_+_42338309 | 0.75 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr23_-_18057851 | 0.75 |
ENSDART00000173075
ENSDART00000173230 ENSDART00000173135 ENSDART00000173431 ENSDART00000173068 ENSDART00000172987 |
zgc:92287
|
zgc:92287 |
| chr6_-_13709591 | 0.74 |
ENSDART00000151771
|
chpfb
|
chondroitin polymerizing factor b |
| chr16_-_47381519 | 0.74 |
ENSDART00000032188
ENSDART00000150136 |
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr15_-_5624361 | 0.73 |
ENSDART00000176446
ENSDART00000114410 |
wdr62
|
WD repeat domain 62 |
| chr21_-_17016788 | 0.72 |
ENSDART00000186703
|
AL844518.1
|
|
| chr11_+_18873619 | 0.71 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr1_-_20593778 | 0.71 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr6_-_12912606 | 0.71 |
ENSDART00000164640
|
ical1
|
islet cell autoantigen 1-like |
| chr23_-_19831739 | 0.70 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr2_+_27855346 | 0.70 |
ENSDART00000175159
ENSDART00000192645 |
buc
|
bucky ball |
| chr7_+_67748939 | 0.69 |
ENSDART00000162978
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr14_-_16810401 | 0.68 |
ENSDART00000158396
ENSDART00000170758 |
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr4_+_13901458 | 0.68 |
ENSDART00000137549
|
pphln1
|
periphilin 1 |
| chr23_+_4741543 | 0.68 |
ENSDART00000144761
|
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a |
| chr7_+_17096281 | 0.68 |
ENSDART00000035558
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr18_+_13248956 | 0.67 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr5_+_62374092 | 0.67 |
ENSDART00000082965
|
CU928117.1
|
|
| chr14_-_45551572 | 0.67 |
ENSDART00000111410
|
ganab
|
glucosidase, alpha; neutral AB |
| chr8_-_14554785 | 0.67 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr5_+_58679071 | 0.66 |
ENSDART00000019561
|
zgc:171734
|
zgc:171734 |
| chr8_-_13735572 | 0.66 |
ENSDART00000139642
|
si:dkey-258f14.7
|
si:dkey-258f14.7 |
| chr13_+_8892784 | 0.66 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr11_+_24313931 | 0.65 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr23_-_25686894 | 0.65 |
ENSDART00000181420
ENSDART00000088208 |
lrp1ab
|
low density lipoprotein receptor-related protein 1Ab |
| chr12_-_3773869 | 0.65 |
ENSDART00000092983
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr22_+_30046365 | 0.65 |
ENSDART00000192546
|
add3a
|
adducin 3 (gamma) a |
| chr8_-_11324143 | 0.64 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr10_-_26218354 | 0.64 |
ENSDART00000180764
|
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr3_-_58165254 | 0.64 |
ENSDART00000093031
|
snu13a
|
SNU13 homolog, small nuclear ribonucleoprotein a (U4/U6.U5) |
| chr23_-_18130264 | 0.63 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr23_-_18057553 | 0.63 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr15_+_29024895 | 0.63 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr17_+_132555 | 0.62 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr2_+_21048661 | 0.62 |
ENSDART00000156876
|
rreb1b
|
ras responsive element binding protein 1b |
| chr16_+_19637384 | 0.61 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr11_-_26701611 | 0.61 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr20_-_40367493 | 0.60 |
ENSDART00000075096
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr17_+_23462972 | 0.59 |
ENSDART00000112959
ENSDART00000192168 |
ankrd1a
|
ankyrin repeat domain 1a (cardiac muscle) |
| chr12_+_33484458 | 0.59 |
ENSDART00000000069
|
slc9a3r1a
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1a |
| chr2_+_34967022 | 0.58 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr4_-_21466480 | 0.58 |
ENSDART00000139962
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr21_+_30673183 | 0.57 |
ENSDART00000144652
|
clcn5a
|
chloride channel, voltage-sensitive 5a |
| chr1_+_59328030 | 0.57 |
ENSDART00000172464
|
CABZ01052576.1
|
|
| chr1_+_594736 | 0.57 |
ENSDART00000166731
|
jam2a
|
junctional adhesion molecule 2a |
| chr5_-_64431927 | 0.56 |
ENSDART00000158248
|
brd3b
|
bromodomain containing 3b |
| chr14_-_47963115 | 0.56 |
ENSDART00000003826
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr6_-_55423220 | 0.55 |
ENSDART00000158929
|
ctsa
|
cathepsin A |
| chr15_-_23485752 | 0.55 |
ENSDART00000152706
|
nlrx1
|
NLR family member X1 |
| chr21_-_34261677 | 0.54 |
ENSDART00000124649
ENSDART00000172381 ENSDART00000064320 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr8_-_15211164 | 0.53 |
ENSDART00000110550
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr2_-_27855576 | 0.52 |
ENSDART00000121623
|
FO834800.1
|
|
| chr2_+_23048620 | 0.52 |
ENSDART00000170669
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr2_-_39086540 | 0.52 |
ENSDART00000110826
|
NMNAT3
|
si:ch73-170l17.1 |
| chr9_-_16140860 | 0.51 |
ENSDART00000142974
|
myo1b
|
myosin IB |
| chr20_-_28404362 | 0.51 |
ENSDART00000055932
ENSDART00000188161 |
pigh
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr20_-_874807 | 0.51 |
ENSDART00000020506
|
snx14
|
sorting nexin 14 |
| chr5_+_31959954 | 0.51 |
ENSDART00000142826
|
myo1hb
|
myosin IHb |
| chr3_+_53240562 | 0.50 |
ENSDART00000031234
|
stxbp2
|
syntaxin binding protein 2 |
| chr20_+_21391181 | 0.50 |
ENSDART00000185158
ENSDART00000049586 ENSDART00000024922 |
jag2b
|
jagged 2b |
| chr22_-_18546649 | 0.50 |
ENSDART00000171277
|
cirbpb
|
cold inducible RNA binding protein b |
| chr2_+_34967210 | 0.49 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr9_-_29578037 | 0.49 |
ENSDART00000189026
|
cenpj
|
centromere protein J |
| chr8_+_44677569 | 0.48 |
ENSDART00000191104
|
grk5l
|
G protein-coupled receptor kinase 5 like |
| chr15_-_34878388 | 0.48 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr6_-_10728921 | 0.48 |
ENSDART00000151484
|
sp3b
|
Sp3b transcription factor |
| chr24_+_33802528 | 0.47 |
ENSDART00000136040
ENSDART00000147499 ENSDART00000182322 |
atg9b
|
autophagy related 9B |
| chr10_-_1961930 | 0.47 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr14_-_32631013 | 0.47 |
ENSDART00000176815
|
atp11c
|
ATPase phospholipid transporting 11C |
| chr16_-_42066523 | 0.47 |
ENSDART00000180538
ENSDART00000058620 |
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr16_+_26601364 | 0.47 |
ENSDART00000087537
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr17_+_15983247 | 0.47 |
ENSDART00000154275
|
clmn
|
calmin |
| chr10_-_14929630 | 0.47 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
| chr20_-_2298970 | 0.46 |
ENSDART00000136067
|
EPB41L2
|
si:ch73-18b11.1 |
| chr18_+_9493720 | 0.46 |
ENSDART00000053125
|
sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr2_-_37280617 | 0.46 |
ENSDART00000190458
|
nadkb
|
NAD kinase b |
| chr17_-_11151655 | 0.46 |
ENSDART00000156383
|
CU179699.1
|
|
| chr23_-_31810391 | 0.46 |
ENSDART00000189749
ENSDART00000180850 |
hbs1l
|
HBS1-like translational GTPase |
| chr6_-_28961660 | 0.45 |
ENSDART00000147285
|
tomm70a
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr3_-_21061931 | 0.45 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr2_+_21000334 | 0.45 |
ENSDART00000062563
ENSDART00000147809 |
rreb1b
|
ras responsive element binding protein 1b |
| chr25_-_893464 | 0.45 |
ENSDART00000159321
|
znf609a
|
zinc finger protein 609a |
| chr2_+_37480669 | 0.44 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr17_-_8692722 | 0.44 |
ENSDART00000148931
ENSDART00000192891 |
ctbp2a
|
C-terminal binding protein 2a |
| chr7_+_38962459 | 0.44 |
ENSDART00000173851
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr5_-_69923241 | 0.43 |
ENSDART00000187389
|
fktn
|
fukutin |
| chr6_-_35032792 | 0.43 |
ENSDART00000168256
|
ddr2a
|
discoidin domain receptor tyrosine kinase 2a |
| chr7_+_52887701 | 0.42 |
ENSDART00000109973
|
tp53bp1
|
tumor protein p53 binding protein, 1 |
| chr8_+_54135642 | 0.42 |
ENSDART00000170712
|
brpf1
|
bromodomain and PHD finger containing, 1 |
| chr21_-_25722834 | 0.41 |
ENSDART00000101208
|
abhd11
|
abhydrolase domain containing 11 |
| chr11_+_31730680 | 0.41 |
ENSDART00000145497
|
diaph3
|
diaphanous-related formin 3 |
| chr23_-_42883522 | 0.39 |
ENSDART00000148500
|
dlgap4a
|
discs, large (Drosophila) homolog-associated protein 4a |
| chr21_-_25522906 | 0.39 |
ENSDART00000110923
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr1_-_50293946 | 0.39 |
ENSDART00000053028
ENSDART00000125099 |
tbck
|
TBC1 domain containing kinase |
| chr10_-_1961576 | 0.38 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr24_-_35707552 | 0.38 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr5_+_20030414 | 0.38 |
ENSDART00000181430
ENSDART00000047841 ENSDART00000182813 |
sgsm1a
|
small G protein signaling modulator 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of figla
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.3 | 3.5 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.3 | 0.9 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.3 | 1.4 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.3 | 1.0 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.2 | 1.5 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.2 | 0.9 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.2 | 1.8 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 2.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 0.9 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.2 | 1.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 1.9 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.7 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 0.6 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.8 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 1.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.9 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.9 | GO:0021744 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.1 | 1.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 1.1 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 0.3 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 2.0 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.9 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 3.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.4 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 1.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 1.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.7 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.5 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 2.8 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 0.4 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 1.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.5 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.4 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.3 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.3 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 | 1.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.9 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.9 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 1.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.2 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.5 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.6 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.4 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.7 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 1.2 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 1.0 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.6 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.2 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.3 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 2.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.4 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 1.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.5 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.5 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.4 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.5 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.3 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.9 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.1 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.6 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.3 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.4 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 0.1 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.7 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 3.2 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.8 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.7 | GO:0051170 | nuclear import(GO:0051170) |
| 0.0 | 1.0 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.1 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 0.9 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 2.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 1.5 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 2.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.4 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 2.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.3 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 3.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 5.7 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 2.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 3.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.7 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.6 | GO:0099572 | postsynaptic specialization(GO:0099572) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.4 | 1.9 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.4 | 2.9 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.3 | 1.4 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.3 | 1.0 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 1.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 0.9 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.7 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 0.9 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 0.6 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 1.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 2.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.7 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 1.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.9 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 2.0 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.0 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.7 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 2.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.0 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 2.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 0.4 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 1.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 2.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.3 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 2.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 4.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.5 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.7 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.8 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.9 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.1 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.2 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 2.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |