Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for etv7
Z-value: 0.81
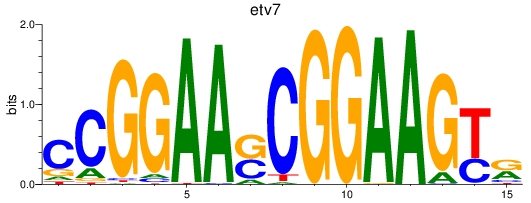
Transcription factors associated with etv7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
etv7
|
ENSDARG00000089434 | ETS variant transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| etv7 | dr11_v1_chr23_-_5101847_5101847 | 0.21 | 4.1e-01 | Click! |
Activity profile of etv7 motif
Sorted Z-values of etv7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_29648854 | 0.95 |
ENSDART00000146280
|
bbip1
|
BBSome interacting protein 1 |
| chr5_-_50084310 | 0.84 |
ENSDART00000074599
ENSDART00000189970 |
fam172a
|
family with sequence similarity 172, member A |
| chr16_+_6944311 | 0.82 |
ENSDART00000144763
|
eaf1
|
ELL associated factor 1 |
| chr5_+_872299 | 0.82 |
ENSDART00000130042
|
fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr19_+_9050852 | 0.77 |
ENSDART00000151031
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr16_-_33930759 | 0.77 |
ENSDART00000177453
|
dnali1
|
dynein, axonemal, light intermediate chain 1 |
| chr21_+_37513058 | 0.76 |
ENSDART00000141096
|
amot
|
angiomotin |
| chr2_-_37134169 | 0.75 |
ENSDART00000146123
ENSDART00000146533 ENSDART00000040427 |
elavl1a
|
ELAV like RNA binding protein 1a |
| chr13_+_9559461 | 0.75 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr19_+_47290287 | 0.75 |
ENSDART00000078382
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr25_-_13659249 | 0.73 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr16_+_6944564 | 0.72 |
ENSDART00000104252
|
eaf1
|
ELL associated factor 1 |
| chr8_-_45430817 | 0.71 |
ENSDART00000150067
ENSDART00000112394 |
ywhabb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide b |
| chr16_-_6944927 | 0.69 |
ENSDART00000149620
|
pmvk
|
phosphomevalonate kinase |
| chr13_-_5978433 | 0.64 |
ENSDART00000102555
|
actr2b
|
ARP2 actin related protein 2b homolog |
| chr3_+_24511959 | 0.64 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr8_-_31053872 | 0.63 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr7_+_29962559 | 0.63 |
ENSDART00000075538
|
fbxo22
|
F-box protein 22 |
| chr12_-_4408828 | 0.63 |
ENSDART00000152447
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr16_+_38337783 | 0.63 |
ENSDART00000135008
|
gabpb2b
|
GA binding protein transcription factor, beta subunit 2b |
| chr25_-_18948816 | 0.62 |
ENSDART00000091549
|
nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr10_+_17747880 | 0.60 |
ENSDART00000135044
|
pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr25_+_16880990 | 0.60 |
ENSDART00000020259
|
zgc:77158
|
zgc:77158 |
| chr22_-_347424 | 0.59 |
ENSDART00000067633
|
necap2
|
NECAP endocytosis associated 2 |
| chr8_+_32389838 | 0.59 |
ENSDART00000076350
ENSDART00000146901 |
mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2 like |
| chr5_+_42467867 | 0.58 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr7_+_46019780 | 0.57 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr8_+_7854130 | 0.56 |
ENSDART00000165575
|
cxxc1a
|
CXXC finger protein 1a |
| chr25_-_37338048 | 0.55 |
ENSDART00000073439
|
trim44
|
tripartite motif containing 44 |
| chr25_-_3627130 | 0.53 |
ENSDART00000171863
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr7_+_756942 | 0.53 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr1_+_494297 | 0.52 |
ENSDART00000108579
ENSDART00000146732 |
blzf1
|
basic leucine zipper nuclear factor 1 |
| chr3_+_62140077 | 0.51 |
ENSDART00000108945
|
GID4
|
GID complex subunit 4 homolog |
| chr21_-_36453594 | 0.51 |
ENSDART00000193176
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr8_-_11546175 | 0.51 |
ENSDART00000081909
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr8_-_33154677 | 0.50 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr16_+_54829293 | 0.50 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr21_+_170038 | 0.50 |
ENSDART00000157614
|
klhl8
|
kelch-like family member 8 |
| chr9_-_7287375 | 0.48 |
ENSDART00000128352
|
mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr21_-_45073764 | 0.48 |
ENSDART00000181390
ENSDART00000063714 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr15_-_26931541 | 0.48 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr14_+_12178915 | 0.48 |
ENSDART00000054626
|
hdac3
|
histone deacetylase 3 |
| chr17_+_43863708 | 0.48 |
ENSDART00000133874
ENSDART00000140316 ENSDART00000142929 ENSDART00000148090 |
zgc:66313
|
zgc:66313 |
| chr16_+_54829574 | 0.47 |
ENSDART00000148392
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr16_+_38338721 | 0.47 |
ENSDART00000076528
ENSDART00000142885 |
gabpb2b
|
GA binding protein transcription factor, beta subunit 2b |
| chr10_+_36439293 | 0.46 |
ENSDART00000043802
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr2_-_24317240 | 0.45 |
ENSDART00000078975
|
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr19_-_37508571 | 0.45 |
ENSDART00000018255
|
ilf2
|
interleukin enhancer binding factor 2 |
| chr10_-_1697037 | 0.44 |
ENSDART00000125188
ENSDART00000002985 |
srsf9
|
serine/arginine-rich splicing factor 9 |
| chr9_+_44304980 | 0.44 |
ENSDART00000147990
|
ssfa2
|
sperm specific antigen 2 |
| chr8_+_4803906 | 0.44 |
ENSDART00000045533
|
tmem127
|
transmembrane protein 127 |
| chr21_-_30254185 | 0.44 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr13_+_49727333 | 0.44 |
ENSDART00000168799
ENSDART00000037559 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr21_-_36453417 | 0.43 |
ENSDART00000018350
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr22_-_506522 | 0.43 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr3_-_25492361 | 0.43 |
ENSDART00000147322
ENSDART00000055473 |
grb2b
|
growth factor receptor-bound protein 2b |
| chr23_+_45229198 | 0.42 |
ENSDART00000172445
|
ttc39b
|
tetratricopeptide repeat domain 39B |
| chr1_+_11107688 | 0.39 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr4_+_2637947 | 0.39 |
ENSDART00000130623
|
dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr5_-_42661012 | 0.39 |
ENSDART00000158339
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr3_+_28831450 | 0.38 |
ENSDART00000055422
|
flr
|
fleer |
| chr11_-_6868287 | 0.38 |
ENSDART00000037824
|
lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr13_+_6188759 | 0.38 |
ENSDART00000161062
|
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr9_-_21825913 | 0.37 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr1_+_54889733 | 0.37 |
ENSDART00000140375
|
zfyve27
|
zinc finger, FYVE domain containing 27 |
| chr6_+_112579 | 0.36 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr13_-_35760969 | 0.36 |
ENSDART00000127476
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr25_+_245438 | 0.36 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr2_+_35854242 | 0.36 |
ENSDART00000134918
|
dhx9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr17_-_39779906 | 0.35 |
ENSDART00000155181
|
pimr61
|
Pim proto-oncogene, serine/threonine kinase, related 61 |
| chr11_-_16975190 | 0.35 |
ENSDART00000122222
|
suclg2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr11_-_6868474 | 0.34 |
ENSDART00000168372
|
lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr7_+_29177191 | 0.34 |
ENSDART00000008096
|
aph1b
|
APH1B gamma secretase subunit |
| chr10_-_41400049 | 0.33 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr22_+_16320076 | 0.33 |
ENSDART00000164161
|
osbpl1a
|
oxysterol binding protein-like 1A |
| chr13_-_35761266 | 0.33 |
ENSDART00000190217
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr13_-_44819254 | 0.32 |
ENSDART00000131673
|
si:dkeyp-2e4.8
|
si:dkeyp-2e4.8 |
| chr2_+_2772447 | 0.31 |
ENSDART00000124882
|
thoc1
|
THO complex 1 |
| chr13_+_6189203 | 0.31 |
ENSDART00000109665
|
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr24_+_33462800 | 0.31 |
ENSDART00000166666
ENSDART00000050826 |
rmc1
|
regulator of MON1-CCZ1 |
| chr3_+_301479 | 0.31 |
ENSDART00000165169
|
CABZ01075611.1
|
|
| chr11_+_5681762 | 0.30 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr8_+_387622 | 0.29 |
ENSDART00000167361
|
pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr14_+_49220026 | 0.29 |
ENSDART00000063643
ENSDART00000128744 |
rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| chr5_+_32490238 | 0.29 |
ENSDART00000191839
|
ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr5_-_38777852 | 0.28 |
ENSDART00000131603
|
si:dkey-58f10.4
|
si:dkey-58f10.4 |
| chr5_-_4297459 | 0.28 |
ENSDART00000018895
|
srrt
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr7_-_33351485 | 0.27 |
ENSDART00000146420
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr12_-_20409794 | 0.27 |
ENSDART00000077936
|
lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr13_+_5978809 | 0.27 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr7_+_26545502 | 0.26 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr6_+_60125033 | 0.25 |
ENSDART00000148557
ENSDART00000008224 |
aurka
|
aurora kinase A |
| chr23_+_32101202 | 0.25 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr23_+_32101361 | 0.25 |
ENSDART00000138849
|
zgc:56699
|
zgc:56699 |
| chr11_-_18449 | 0.25 |
ENSDART00000172050
|
prr13
|
proline rich 13 |
| chr19_-_32888758 | 0.24 |
ENSDART00000052080
|
laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr3_-_8388344 | 0.24 |
ENSDART00000146856
|
rbfox3b
|
RNA binding fox-1 homolog 3b |
| chr1_-_494280 | 0.24 |
ENSDART00000144406
|
ercc5
|
excision repair cross-complementation group 5 |
| chr5_-_68495967 | 0.24 |
ENSDART00000188107
|
ephb4a
|
eph receptor B4a |
| chr1_-_6225285 | 0.24 |
ENSDART00000141653
|
mdh1b
|
malate dehydrogenase 1B, NAD (soluble) |
| chr18_-_40884087 | 0.24 |
ENSDART00000059194
|
snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr7_+_26545911 | 0.24 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr17_+_23937262 | 0.23 |
ENSDART00000113276
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr8_+_52530889 | 0.23 |
ENSDART00000127729
ENSDART00000170360 ENSDART00000162687 |
stambpb
|
STAM binding protein b |
| chr2_+_24507597 | 0.22 |
ENSDART00000133109
|
rps28
|
ribosomal protein S28 |
| chr20_+_27093042 | 0.22 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr1_+_6225493 | 0.21 |
ENSDART00000145378
|
fastkd2
|
FAST kinase domains 2 |
| chr18_+_15778110 | 0.21 |
ENSDART00000014188
|
ube2na
|
ubiquitin-conjugating enzyme E2Na |
| chr11_-_141592 | 0.21 |
ENSDART00000092787
|
cdk4
|
cyclin-dependent kinase 4 |
| chr6_+_18367388 | 0.21 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr19_+_27479563 | 0.21 |
ENSDART00000049368
ENSDART00000185426 |
atat1
|
alpha tubulin acetyltransferase 1 |
| chr20_-_4793450 | 0.20 |
ENSDART00000053870
|
galca
|
galactosylceramidase a |
| chr25_-_3230187 | 0.20 |
ENSDART00000160545
|
cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr11_-_18254 | 0.20 |
ENSDART00000167814
|
prr13
|
proline rich 13 |
| chr14_+_23668730 | 0.20 |
ENSDART00000157741
|
med12
|
mediator complex subunit 12 |
| chr5_+_3927989 | 0.19 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr13_-_28263856 | 0.19 |
ENSDART00000041036
ENSDART00000079806 |
smap1
|
small ArfGAP 1 |
| chr9_-_29003245 | 0.18 |
ENSDART00000183391
ENSDART00000188836 |
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr13_-_3324764 | 0.18 |
ENSDART00000102748
ENSDART00000114040 |
ubr2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr21_-_44556148 | 0.17 |
ENSDART00000163955
|
brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr21_-_13123176 | 0.17 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr6_-_19381993 | 0.17 |
ENSDART00000164114
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr18_+_50707179 | 0.16 |
ENSDART00000160206
|
rap2b
|
RAP2B, member of RAS oncogene family |
| chr22_+_396840 | 0.16 |
ENSDART00000163198
|
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr19_-_10395683 | 0.16 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr17_+_9310259 | 0.16 |
ENSDART00000186158
ENSDART00000190329 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr1_-_354115 | 0.16 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr1_+_22002649 | 0.16 |
ENSDART00000141317
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr15_-_1941042 | 0.15 |
ENSDART00000112946
ENSDART00000155504 |
dock10
|
dedicator of cytokinesis 10 |
| chr22_-_6194517 | 0.15 |
ENSDART00000134757
ENSDART00000181598 ENSDART00000129829 |
si:rp71-36a1.5
|
si:rp71-36a1.5 |
| chr22_+_9862243 | 0.15 |
ENSDART00000105942
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr21_+_21621042 | 0.15 |
ENSDART00000134907
|
tgfb1b
|
transforming growth factor, beta 1b |
| chr2_+_24507770 | 0.14 |
ENSDART00000154802
ENSDART00000052063 |
rps28
|
ribosomal protein S28 |
| chr19_+_63567 | 0.14 |
ENSDART00000165657
ENSDART00000165183 |
zhx2b
|
zinc fingers and homeoboxes 2b |
| chr10_-_105100 | 0.13 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr19_+_27479838 | 0.13 |
ENSDART00000103922
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr22_+_94692 | 0.13 |
ENSDART00000114777
|
nckipsd
|
NCK interacting protein with SH3 domain |
| chr9_-_21067971 | 0.12 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr2_-_58142854 | 0.12 |
ENSDART00000169909
|
CABZ01110881.1
|
|
| chr14_-_38878356 | 0.11 |
ENSDART00000173082
ENSDART00000039788 |
uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr6_-_31364475 | 0.11 |
ENSDART00000145715
ENSDART00000134370 |
ak4
|
adenylate kinase 4 |
| chr2_-_19520324 | 0.11 |
ENSDART00000079877
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr16_+_53489676 | 0.10 |
ENSDART00000074653
|
grinab
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1b (glutamate binding) |
| chr25_+_469855 | 0.10 |
ENSDART00000104717
|
rsl24d1
|
ribosomal L24 domain containing 1 |
| chr4_+_9612574 | 0.10 |
ENSDART00000150336
ENSDART00000041289 ENSDART00000150828 |
tmem243b
|
transmembrane protein 243, mitochondrial b |
| chr21_-_44556411 | 0.09 |
ENSDART00000163983
|
brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr1_-_46505310 | 0.09 |
ENSDART00000178072
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr5_+_483965 | 0.09 |
ENSDART00000150007
|
tek
|
TEK tyrosine kinase, endothelial |
| chr13_-_4979029 | 0.09 |
ENSDART00000132931
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr12_-_46315898 | 0.09 |
ENSDART00000153138
|
si:ch211-226h7.3
|
si:ch211-226h7.3 |
| chr7_+_26762958 | 0.08 |
ENSDART00000167956
ENSDART00000134717 |
tspan18a
|
tetraspanin 18a |
| chr14_+_49251331 | 0.08 |
ENSDART00000148882
|
anxa6
|
annexin A6 |
| chr22_+_9523479 | 0.08 |
ENSDART00000189473
ENSDART00000143953 |
strip1
|
striatin interacting protein 1 |
| chr13_-_33700461 | 0.07 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr8_-_17184482 | 0.07 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr17_-_43863700 | 0.06 |
ENSDART00000157530
|
ahsa1b
|
AHA1, activator of heat shock protein ATPase homolog 1b |
| chr22_+_9862466 | 0.05 |
ENSDART00000146864
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr7_+_44608224 | 0.05 |
ENSDART00000005033
|
cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr6_-_30859656 | 0.05 |
ENSDART00000156235
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr6_+_49722970 | 0.05 |
ENSDART00000155934
ENSDART00000154738 |
stx16
|
syntaxin 16 |
| chr21_+_19858627 | 0.05 |
ENSDART00000147010
|
fybb
|
FYN binding protein b |
| chr1_+_54766943 | 0.05 |
ENSDART00000144759
|
nlrc6
|
NLR family CARD domain containing 6 |
| chr10_+_334627 | 0.05 |
ENSDART00000169455
|
OVOL3
|
zgc:171929 |
| chr2_+_58008980 | 0.05 |
ENSDART00000171264
|
si:ch211-155e24.3
|
si:ch211-155e24.3 |
| chr23_-_45318760 | 0.04 |
ENSDART00000166883
|
ccdc171
|
coiled-coil domain containing 171 |
| chr16_-_45001842 | 0.04 |
ENSDART00000037797
|
sult2st3
|
sulfotransferase family 2, cytosolic sulfotransferase 3 |
| chr1_+_12049229 | 0.04 |
ENSDART00000103403
ENSDART00000137697 |
saraf
|
store-operated calcium entry-associated regulatory factor |
| chr22_+_16535575 | 0.04 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr4_+_29773917 | 0.04 |
ENSDART00000170798
|
BX530097.1
|
|
| chr7_+_44608478 | 0.04 |
ENSDART00000149981
|
cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr9_-_21067673 | 0.03 |
ENSDART00000180257
|
tbx15
|
T-box 15 |
| chr3_-_15444396 | 0.03 |
ENSDART00000104361
|
si:dkey-56d12.4
|
si:dkey-56d12.4 |
| chr21_+_27513859 | 0.03 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr21_+_33454147 | 0.03 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr6_+_49723289 | 0.03 |
ENSDART00000190452
|
stx16
|
syntaxin 16 |
| chr21_-_18932761 | 0.02 |
ENSDART00000140129
|
med15
|
mediator complex subunit 15 |
| chr1_+_961607 | 0.02 |
ENSDART00000184660
|
n6amt1
|
N-6 adenine-specific DNA methyltransferase 1 |
| chr21_-_22928214 | 0.02 |
ENSDART00000182760
|
dub
|
duboraya |
| chr23_+_20644511 | 0.01 |
ENSDART00000133131
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr1_-_56213723 | 0.01 |
ENSDART00000142505
ENSDART00000137237 |
si:dkey-76b14.2
|
si:dkey-76b14.2 |
| chr6_-_55958705 | 0.01 |
ENSDART00000155963
|
eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr7_-_50272912 | 0.01 |
ENSDART00000098842
|
hsd17b12b
|
hydroxysteroid (17-beta) dehydrogenase 12b |
| chr3_+_36972298 | 0.00 |
ENSDART00000150917
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr5_-_45894802 | 0.00 |
ENSDART00000097648
|
crfb6
|
cytokine receptor family member b6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of etv7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.2 | 0.7 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.8 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.6 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.6 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.8 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 0.5 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.3 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.6 | GO:2001032 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 1.5 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.1 | 0.6 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 0.4 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.5 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.2 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.4 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.7 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.6 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.2 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.3 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.0 | 0.1 | GO:0003413 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.0 | 0.4 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.8 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.4 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.6 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.4 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.1 | 1.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.3 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.4 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.7 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.1 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.8 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0005771 | multivesicular body(GO:0005771) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 0.7 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.2 | 0.8 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.6 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.4 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 0.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.3 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.6 | GO:0015154 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.6 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.7 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.5 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.3 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.8 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.9 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |