Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
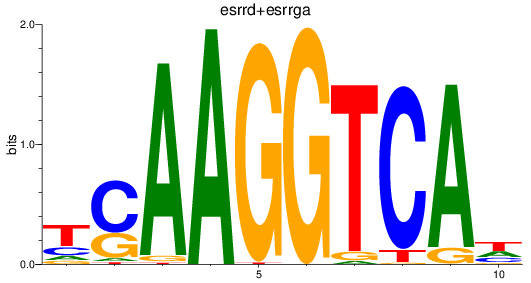
Results for esrrd+esrrga_esrrb_esrra+esrrgb
Z-value: 2.49
Transcription factors associated with esrrd+esrrga_esrrb_esrra+esrrgb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
esrrga
|
ENSDARG00000004861 | estrogen-related receptor gamma a |
|
esrrd
|
ENSDARG00000015064 | estrogen-related receptor delta |
|
esrrb
|
ENSDARG00000100847 | estrogen-related receptor beta |
|
esrrgb
|
ENSDARG00000011696 | estrogen-related receptor gamma b |
|
esrra
|
ENSDARG00000069266 | estrogen-related receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| esrrgb | dr11_v1_chr20_-_46808870_46808870 | -0.80 | 6.3e-05 | Click! |
| esrrb | dr11_v1_chr17_-_29902187_29902187 | 0.77 | 1.6e-04 | Click! |
| esrrd | dr11_v1_chr18_-_48992363_48992363 | 0.73 | 5.2e-04 | Click! |
| esrra | dr11_v1_chr21_+_26620867_26620867 | 0.35 | 1.6e-01 | Click! |
Activity profile of esrrd+esrrga_esrrb_esrra+esrrgb motif
Sorted Z-values of esrrd+esrrga_esrrb_esrra+esrrgb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_39568290 | 8.69 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr22_+_25720725 | 8.24 |
ENSDART00000150778
|
si:dkeyp-98a7.8
|
si:dkeyp-98a7.8 |
| chr16_-_17197546 | 7.02 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr17_-_26610814 | 6.05 |
ENSDART00000133402
ENSDART00000016608 |
mrpl57
|
mitochondrial ribosomal protein L57 |
| chr16_-_46578523 | 5.99 |
ENSDART00000131061
|
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr8_-_52715911 | 5.95 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr22_+_26853254 | 5.82 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr21_+_5080789 | 5.11 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr3_+_1182315 | 5.08 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr23_+_44611864 | 5.05 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr12_-_25201576 | 4.68 |
ENSDART00000077188
|
cox7a3
|
cytochrome c oxidase subunit VIIa polypeptide 3 |
| chr11_+_18175893 | 4.67 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr18_-_39636348 | 4.55 |
ENSDART00000129828
|
cyp19a1a
|
cytochrome P450, family 19, subfamily A, polypeptide 1a |
| chr6_-_10835849 | 4.43 |
ENSDART00000005903
ENSDART00000135065 |
atp5mc3b
|
ATP synthase membrane subunit c locus 3b |
| chr7_+_38716048 | 4.43 |
ENSDART00000024590
|
syt13
|
synaptotagmin XIII |
| chr21_+_37411547 | 4.32 |
ENSDART00000076320
|
mrps17
|
mitochondrial ribosomal protein S17 |
| chr7_+_22293894 | 4.14 |
ENSDART00000056790
|
tmem256
|
transmembrane protein 256 |
| chr11_+_18037729 | 3.99 |
ENSDART00000111624
|
zgc:175135
|
zgc:175135 |
| chr5_-_57820873 | 3.93 |
ENSDART00000089961
|
sik2a
|
salt-inducible kinase 2a |
| chr11_+_18053333 | 3.87 |
ENSDART00000075750
|
zgc:175135
|
zgc:175135 |
| chr8_+_39760258 | 3.78 |
ENSDART00000037914
|
cox6a1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr14_+_32852388 | 3.75 |
ENSDART00000166351
|
nkrf
|
NFKB repressing factor |
| chr16_-_4546965 | 3.62 |
ENSDART00000060594
|
mrpl51
|
mitochondrial ribosomal protein L51 |
| chr3_+_24207243 | 3.55 |
ENSDART00000023454
ENSDART00000136400 |
adsl
|
adenylosuccinate lyase |
| chr6_-_33878665 | 3.47 |
ENSDART00000129916
|
tmem69
|
transmembrane protein 69 |
| chr3_-_13599482 | 3.46 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr3_-_34724879 | 3.45 |
ENSDART00000177021
|
thraa
|
thyroid hormone receptor alpha a |
| chr6_+_52235441 | 3.41 |
ENSDART00000056319
|
cox6c
|
cytochrome c oxidase subunit VIc |
| chr16_-_4547193 | 3.38 |
ENSDART00000097540
|
mrpl51
|
mitochondrial ribosomal protein L51 |
| chr5_-_1076431 | 3.38 |
ENSDART00000168336
|
si:zfos-128g4.1
|
si:zfos-128g4.1 |
| chr11_+_18130300 | 3.35 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr21_-_22681534 | 3.33 |
ENSDART00000159233
|
gig2f
|
grass carp reovirus (GCRV)-induced gene 2f |
| chr11_+_18157260 | 3.30 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr3_+_3454610 | 3.28 |
ENSDART00000024900
|
zgc:165453
|
zgc:165453 |
| chr3_+_58167288 | 3.27 |
ENSDART00000155874
ENSDART00000010395 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein 2a |
| chr3_-_60142530 | 3.24 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr1_+_580642 | 3.06 |
ENSDART00000147633
|
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr20_-_16171297 | 2.83 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr5_-_57879138 | 2.81 |
ENSDART00000145959
|
sik2a
|
salt-inducible kinase 2a |
| chr21_+_39197628 | 2.80 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr16_-_5721386 | 2.72 |
ENSDART00000136655
|
ndufa3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 |
| chr12_-_47648538 | 2.58 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr14_-_35892767 | 2.52 |
ENSDART00000052648
|
tmem144b
|
transmembrane protein 144b |
| chr18_+_30508729 | 2.48 |
ENSDART00000185140
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr16_-_50229193 | 2.48 |
ENSDART00000161782
ENSDART00000010081 |
etfb
|
electron-transfer-flavoprotein, beta polypeptide |
| chr10_-_16065185 | 2.42 |
ENSDART00000187266
|
si:dkey-184a18.5
|
si:dkey-184a18.5 |
| chr13_-_18195942 | 2.34 |
ENSDART00000079902
|
slc25a16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr24_-_23974559 | 2.29 |
ENSDART00000080510
ENSDART00000135242 |
ndufb4
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4 |
| chr12_+_48395693 | 2.25 |
ENSDART00000180362
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr22_-_21150845 | 2.24 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr6_+_9952678 | 2.23 |
ENSDART00000019325
|
cyp20a1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr6_+_22337081 | 2.20 |
ENSDART00000128047
ENSDART00000138930 |
uqcrc1
|
ubiquinol-cytochrome c reductase core protein 1 |
| chr1_+_24557414 | 2.11 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr21_-_28640316 | 2.08 |
ENSDART00000128237
|
nrg2a
|
neuregulin 2a |
| chr5_-_37886063 | 2.08 |
ENSDART00000131378
ENSDART00000132152 |
si:ch211-139a5.9
|
si:ch211-139a5.9 |
| chr8_+_45338073 | 2.04 |
ENSDART00000185024
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr21_+_27336550 | 2.02 |
ENSDART00000139634
|
dpp3
|
dipeptidyl-peptidase 3 |
| chr13_-_36418921 | 2.02 |
ENSDART00000135804
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr22_-_26865361 | 2.00 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr19_-_5103141 | 1.99 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr7_-_28549361 | 1.96 |
ENSDART00000173918
ENSDART00000054368 ENSDART00000113313 |
st5
|
suppression of tumorigenicity 5 |
| chr7_+_48805534 | 1.92 |
ENSDART00000145375
ENSDART00000148744 |
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr23_+_19198244 | 1.88 |
ENSDART00000047015
|
ccdc115
|
coiled-coil domain containing 115 |
| chr11_+_13207898 | 1.87 |
ENSDART00000060310
|
atp5f1b
|
ATP synthase F1 subunit beta |
| chr5_+_37729207 | 1.84 |
ENSDART00000184378
|
cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr18_+_30507839 | 1.83 |
ENSDART00000026866
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr7_+_48805725 | 1.81 |
ENSDART00000166543
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr10_+_7671260 | 1.80 |
ENSDART00000157608
|
fam136a
|
family with sequence similarity 136, member A |
| chr19_+_1465004 | 1.76 |
ENSDART00000159157
|
CABZ01073736.1
|
|
| chr25_+_16116740 | 1.76 |
ENSDART00000139778
|
far1
|
fatty acyl CoA reductase 1 |
| chr19_-_5103313 | 1.75 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr21_+_15704556 | 1.73 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr13_-_15082024 | 1.72 |
ENSDART00000157482
|
sfxn5a
|
sideroflexin 5a |
| chr12_-_45238759 | 1.70 |
ENSDART00000154859
|
trim65
|
tripartite motif containing 65 |
| chr5_+_61738276 | 1.70 |
ENSDART00000186256
|
RASL10B
|
RAS like family 10 member B |
| chr19_-_7291733 | 1.64 |
ENSDART00000015559
|
sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr2_+_30249977 | 1.63 |
ENSDART00000109160
ENSDART00000135171 |
tmem70
|
transmembrane protein 70 |
| chr22_-_15720347 | 1.63 |
ENSDART00000105692
|
mrpl54
|
mitochondrial ribosomal protein L54 |
| chr17_-_38778826 | 1.58 |
ENSDART00000168182
ENSDART00000124041 ENSDART00000136921 |
dglucy
|
D-glutamate cyclase |
| chr22_-_7050 | 1.56 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr12_+_28799988 | 1.55 |
ENSDART00000022724
|
pnpo
|
pyridoxamine 5'-phosphate oxidase |
| chr1_-_55262763 | 1.55 |
ENSDART00000152769
|
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr13_-_9311253 | 1.54 |
ENSDART00000058056
|
mrps26
|
mitochondrial ribosomal protein S26 |
| chr22_+_24215007 | 1.53 |
ENSDART00000162227
|
glrx2
|
glutaredoxin 2 |
| chr1_+_12335816 | 1.53 |
ENSDART00000067086
|
nansa
|
N-acetylneuraminic acid synthase a |
| chr21_+_5882300 | 1.52 |
ENSDART00000165065
|
uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr1_-_55248496 | 1.51 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr21_+_11885404 | 1.51 |
ENSDART00000092015
|
dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr9_-_27717006 | 1.49 |
ENSDART00000146860
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr13_+_15800742 | 1.48 |
ENSDART00000146234
|
apopt1
|
apoptogenic 1, mitochondrial |
| chr16_+_33938227 | 1.46 |
ENSDART00000166254
|
gpn2
|
GPN-loop GTPase 2 |
| chr17_-_1703259 | 1.45 |
ENSDART00000156489
|
xgb
|
x globin |
| chr10_-_18463934 | 1.43 |
ENSDART00000133116
ENSDART00000113422 |
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr8_-_49495584 | 1.40 |
ENSDART00000141691
|
opn7d
|
opsin 7, group member d |
| chr2_+_44545912 | 1.40 |
ENSDART00000155362
|
si:dkeyp-94h10.5
|
si:dkeyp-94h10.5 |
| chr15_-_21692630 | 1.39 |
ENSDART00000039865
|
sdhdb
|
succinate dehydrogenase complex, subunit D, integral membrane protein b |
| chr2_-_1227221 | 1.39 |
ENSDART00000130897
|
ABCF3
|
ATP binding cassette subfamily F member 3 |
| chr14_-_17575764 | 1.38 |
ENSDART00000123145
|
rnf4
|
ring finger protein 4 |
| chr1_+_26467071 | 1.35 |
ENSDART00000112329
ENSDART00000159318 ENSDART00000193833 ENSDART00000011809 |
uso1
|
USO1 vesicle transport factor |
| chr5_+_50879545 | 1.33 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr17_+_15213496 | 1.33 |
ENSDART00000058351
ENSDART00000131663 |
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr21_+_39432248 | 1.32 |
ENSDART00000179938
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr9_+_7909490 | 1.31 |
ENSDART00000133063
ENSDART00000109288 |
myo16
|
myosin XVI |
| chr14_+_1240419 | 1.31 |
ENSDART00000181248
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr2_-_56649883 | 1.31 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr15_-_1484795 | 1.30 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr14_-_35414559 | 1.29 |
ENSDART00000145033
|
rnaseh2c
|
ribonuclease H2, subunit C |
| chr11_+_14057605 | 1.28 |
ENSDART00000166262
|
grin3bb
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3Bb |
| chr14_+_1240235 | 1.27 |
ENSDART00000127477
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr13_-_330004 | 1.26 |
ENSDART00000093149
|
ddx21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr7_-_41915312 | 1.26 |
ENSDART00000159869
|
dnaja2
|
DnaJ (Hsp40) homolog, subfamily A, member 2 |
| chr6_+_50381665 | 1.25 |
ENSDART00000141128
|
cyc1
|
cytochrome c-1 |
| chr9_-_29844596 | 1.24 |
ENSDART00000138574
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr21_-_5879897 | 1.23 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr23_-_20002459 | 1.23 |
ENSDART00000163396
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr7_+_40094081 | 1.21 |
ENSDART00000186054
|
si:ch73-174h16.4
|
si:ch73-174h16.4 |
| chr3_-_32337653 | 1.21 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr22_+_33362552 | 1.20 |
ENSDART00000101580
|
nicn1
|
nicolin 1 |
| chr10_+_45128375 | 1.19 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr19_-_44065581 | 1.19 |
ENSDART00000006338
|
mterf3
|
mitochondrial transcription termination factor 3 |
| chr19_+_20163826 | 1.16 |
ENSDART00000090942
ENSDART00000134650 |
ccdc126
|
coiled-coil domain containing 126 |
| chr15_-_1485086 | 1.15 |
ENSDART00000191651
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr19_+_28187480 | 1.14 |
ENSDART00000183825
|
irx4b
|
iroquois homeobox 4b |
| chr8_+_37749263 | 1.13 |
ENSDART00000108556
ENSDART00000147942 |
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
| chr15_+_34592215 | 1.13 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr3_-_32590164 | 1.12 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr1_-_46862190 | 1.12 |
ENSDART00000145167
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr22_-_9860792 | 1.08 |
ENSDART00000155908
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr15_+_20801253 | 1.07 |
ENSDART00000179387
|
aldh3a1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr13_-_12660318 | 1.06 |
ENSDART00000008498
|
adh8a
|
alcohol dehydrogenase 8a |
| chr25_+_25124684 | 1.04 |
ENSDART00000167542
|
ldha
|
lactate dehydrogenase A4 |
| chr13_-_38039871 | 1.04 |
ENSDART00000140645
|
CR456624.1
|
|
| chr22_-_9861531 | 1.03 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr24_+_26140855 | 1.02 |
ENSDART00000139017
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr5_-_20491198 | 0.99 |
ENSDART00000183051
ENSDART00000144232 |
ficd
|
FIC domain containing |
| chr15_-_25365570 | 0.99 |
ENSDART00000152754
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr14_+_38878482 | 0.99 |
ENSDART00000043317
|
timm8a
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr14_+_1240604 | 0.98 |
ENSDART00000188258
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr23_+_9560991 | 0.96 |
ENSDART00000081433
ENSDART00000131594 ENSDART00000130069 ENSDART00000138601 |
adrm1
|
adhesion regulating molecule 1 |
| chr6_+_59944488 | 0.96 |
ENSDART00000161158
|
nufip1
|
nuclear fragile X mental retardation protein interacting protein 1 |
| chr11_-_45429199 | 0.94 |
ENSDART00000173111
|
rfc4
|
replication factor C (activator 1) 4 |
| chr6_+_50381347 | 0.94 |
ENSDART00000055504
|
cyc1
|
cytochrome c-1 |
| chr7_-_8344243 | 0.93 |
ENSDART00000149510
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr23_+_9560797 | 0.93 |
ENSDART00000180014
|
adrm1
|
adhesion regulating molecule 1 |
| chr10_-_32558917 | 0.92 |
ENSDART00000128888
ENSDART00000143301 |
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr17_-_33289304 | 0.92 |
ENSDART00000135118
ENSDART00000040346 |
efr3ba
|
EFR3 homolog Ba (S. cerevisiae) |
| chr1_-_17569793 | 0.92 |
ENSDART00000125125
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr7_+_21275152 | 0.91 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr3_+_26814030 | 0.90 |
ENSDART00000180128
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr8_+_38564942 | 0.90 |
ENSDART00000085371
|
rfk
|
riboflavin kinase |
| chr25_+_17870799 | 0.90 |
ENSDART00000136797
|
btbd10a
|
BTB (POZ) domain containing 10a |
| chr16_-_12512568 | 0.89 |
ENSDART00000055161
ENSDART00000160906 |
cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr13_-_18105278 | 0.89 |
ENSDART00000090475
|
washc2c
|
WASH complex subunit 2C |
| chr24_-_6024466 | 0.89 |
ENSDART00000040865
|
pdss1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr9_+_31075896 | 0.88 |
ENSDART00000188042
|
clybl
|
citrate lyase beta like |
| chr3_+_39566999 | 0.87 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr20_-_3290791 | 0.87 |
ENSDART00000146870
|
si:ch1073-412h12.3
|
si:ch1073-412h12.3 |
| chr5_-_26566435 | 0.87 |
ENSDART00000146070
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr6_+_29707129 | 0.87 |
ENSDART00000023549
ENSDART00000164849 |
phb2b
|
prohibitin 2b |
| chr19_+_19976990 | 0.87 |
ENSDART00000052627
|
npvf
|
neuropeptide VF precursor |
| chr7_+_20524064 | 0.86 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr10_-_7671219 | 0.85 |
ENSDART00000159330
|
pcyox1
|
prenylcysteine oxidase 1 |
| chr23_+_2906031 | 0.85 |
ENSDART00000109304
|
c23h20orf24
|
c23h20orf24 homolog (H. sapiens) |
| chr2_-_44971551 | 0.85 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr12_-_290782 | 0.84 |
ENSDART00000152527
ENSDART00000105694 |
adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr5_+_69780493 | 0.84 |
ENSDART00000154507
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr20_+_43942278 | 0.84 |
ENSDART00000100571
|
clic5b
|
chloride intracellular channel 5b |
| chr2_+_26060528 | 0.84 |
ENSDART00000058111
|
grin3ba
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3Ba |
| chr22_+_10201826 | 0.84 |
ENSDART00000006513
ENSDART00000132641 |
pdhb
|
pyruvate dehydrogenase E1 beta subunit |
| chr9_+_27379014 | 0.83 |
ENSDART00000167366
|
tlr20.4
|
toll-like receptor 20, tandem duplicate 4 |
| chr7_-_19600181 | 0.83 |
ENSDART00000100757
|
oxa1l
|
oxidase (cytochrome c) assembly 1-like |
| chr11_+_15890984 | 0.82 |
ENSDART00000158433
|
pank4
|
pantothenate kinase 4 |
| chr23_+_42482137 | 0.82 |
ENSDART00000160199
|
cyp2aa3
|
cytochrome P450, family 2, subfamily AA, polypeptide 3 |
| chr20_-_40758410 | 0.81 |
ENSDART00000183031
|
cx34.5
|
connexin 34.5 |
| chr10_-_57270 | 0.81 |
ENSDART00000058411
|
hlcs
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
| chr1_+_51221320 | 0.81 |
ENSDART00000138878
|
ndufaf5
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 5 |
| chr18_+_44769027 | 0.81 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr18_+_910992 | 0.80 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr25_+_36311333 | 0.80 |
ENSDART00000190174
|
hist1h2a2
|
histone cluster 1 H2A family member 2 |
| chr16_+_38240027 | 0.78 |
ENSDART00000111081
|
prune
|
prune exopolyphosphatase |
| chr16_+_20871021 | 0.77 |
ENSDART00000006429
|
hibadhb
|
3-hydroxyisobutyrate dehydrogenase b |
| chr11_-_11471857 | 0.77 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr4_-_6806632 | 0.77 |
ENSDART00000139242
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr25_-_35139520 | 0.76 |
ENSDART00000189008
|
CR762436.1
|
|
| chr10_+_2234283 | 0.76 |
ENSDART00000136363
|
cntnap3
|
contactin associated protein like 3 |
| chr2_+_10006839 | 0.75 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr21_+_35328025 | 0.74 |
ENSDART00000136211
|
rars
|
arginyl-tRNA synthetase |
| chr21_-_25250594 | 0.74 |
ENSDART00000163862
|
nfrkb
|
nuclear factor related to kappaB binding protein |
| chr14_-_28568107 | 0.71 |
ENSDART00000042850
ENSDART00000145502 |
insb
|
preproinsulin b |
| chr8_+_17184602 | 0.71 |
ENSDART00000050228
ENSDART00000140531 |
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr2_+_30032303 | 0.71 |
ENSDART00000151841
|
rbm33b
|
RNA binding motif protein 33b |
| chr13_-_44836727 | 0.71 |
ENSDART00000144385
|
si:dkeyp-2e4.3
|
si:dkeyp-2e4.3 |
| chr6_+_19383267 | 0.70 |
ENSDART00000166549
|
mchr1a
|
melanin-concentrating hormone receptor 1a |
| chr24_-_21471389 | 0.70 |
ENSDART00000109848
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr5_+_32924669 | 0.69 |
ENSDART00000085219
|
lmo4a
|
LIM domain only 4a |
| chr7_+_17120811 | 0.69 |
ENSDART00000147140
|
prmt3
|
protein arginine methyltransferase 3 |
| chr2_-_3045861 | 0.69 |
ENSDART00000105818
ENSDART00000187575 |
guk1a
|
guanylate kinase 1a |
| chr20_+_16170848 | 0.69 |
ENSDART00000182115
|
zyg11
|
zyg-11 homolog (C. elegans) |
| chr23_-_45568816 | 0.69 |
ENSDART00000076086
|
cyp17a2
|
cytochrome P450, family 17, subfamily A, polypeptide 2 |
| chr25_+_22730490 | 0.69 |
ENSDART00000149455
|
abcc8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of esrrd+esrrga_esrrb_esrra+esrrgb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0030238 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 1.3 | 3.8 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.9 | 3.7 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.8 | 2.4 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.8 | 2.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.7 | 2.1 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.6 | 2.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.6 | 5.0 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.6 | 1.9 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.6 | 3.0 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.6 | 2.3 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.5 | 1.5 | GO:0001113 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.5 | 12.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.4 | 1.3 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.4 | 9.6 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.4 | 4.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.4 | 5.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 3.5 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.4 | 1.5 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.4 | 3.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.3 | 3.4 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.3 | 1.4 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.3 | 2.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.3 | 1.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 1.0 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.3 | 2.1 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.3 | 1.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.3 | 3.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.3 | 1.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 2.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.9 | GO:0045938 | regulation of endocrine process(GO:0044060) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) endocrine hormone secretion(GO:0060986) |
| 0.2 | 0.8 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.2 | 1.4 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.8 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 6.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.2 | 6.4 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.2 | 1.4 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 2.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 1.8 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.2 | 1.0 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 0.8 | GO:0030328 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.2 | 0.8 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 0.5 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.2 | 8.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 1.1 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.2 | 0.9 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.9 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.5 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.5 | GO:0051645 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.1 | 1.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.4 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 1.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.7 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.1 | 0.3 | GO:0009750 | response to fructose(GO:0009750) |
| 0.1 | 5.0 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 1.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.3 | GO:0097676 | histone H3-K4 trimethylation(GO:0080182) histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 0.2 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.1 | 2.0 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.1 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.4 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 1.2 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.1 | 1.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.6 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.8 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 0.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.4 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 1.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 1.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 8.9 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.1 | 7.2 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.1 | 2.4 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.3 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 2.7 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 0.7 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.5 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 1.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.2 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.1 | 1.3 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.1 | 0.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 1.8 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 0.2 | GO:0002637 | regulation of immunoglobulin production(GO:0002637) positive regulation of immunoglobulin production(GO:0002639) |
| 0.1 | 0.8 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 0.8 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.2 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.2 | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.8 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.1 | 0.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.7 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:1903059 | natural killer cell activation(GO:0030101) regulation of lipoprotein metabolic process(GO:0050746) regulation of protein lipidation(GO:1903059) |
| 0.0 | 0.3 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.0 | 0.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.3 | GO:0003272 | endocardial cushion formation(GO:0003272) protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 1.6 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.8 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.7 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.4 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.5 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.7 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0099625 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell action potential(GO:0086005) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.1 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 3.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.7 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.1 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.2 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.2 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.4 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0003250 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.1 | GO:1901492 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 1.5 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.6 | GO:0019319 | hexose biosynthetic process(GO:0019319) monosaccharide biosynthetic process(GO:0046364) |
| 0.0 | 0.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.2 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.6 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 1.3 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.2 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.6 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.4 | GO:0009749 | response to glucose(GO:0009749) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.2 | GO:0001840 | neural plate development(GO:0001840) |
| 0.0 | 1.2 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.4 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.2 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.9 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 0.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.2 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.5 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.4 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 1.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 1.2 | GO:0016051 | carbohydrate biosynthetic process(GO:0016051) |
| 0.0 | 0.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.1 | GO:0046461 | neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 0.0 | 0.0 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) |
| 0.0 | 1.0 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.8 | GO:0001666 | response to hypoxia(GO:0001666) response to decreased oxygen levels(GO:0036293) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.4 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.6 | 8.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.6 | 5.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.6 | 3.0 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.4 | 1.3 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.4 | 3.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.3 | 2.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 1.3 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.3 | 1.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 21.6 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.3 | 4.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 1.5 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.3 | 0.8 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.2 | 7.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 1.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 1.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 3.0 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 5.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 2.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.9 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 3.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.5 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 5.5 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 0.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.7 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 4.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 1.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.0 | 1.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 5.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.7 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 8.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 4.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 8.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 18.6 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 5.6 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.2 | GO:0098858 | actin-based cell projection(GO:0098858) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 2.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 14.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.3 | 5.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 1.2 | 9.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.9 | 3.7 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.9 | 3.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.8 | 4.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.6 | 5.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.6 | 4.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.6 | 2.3 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.5 | 1.6 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.5 | 8.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.5 | 2.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.4 | 1.2 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.4 | 2.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.3 | 2.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 3.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.3 | 1.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 1.5 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.3 | 0.9 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.3 | 1.7 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.3 | 7.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 0.8 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.3 | 1.8 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 1.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 0.7 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.2 | 2.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.7 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.2 | 0.9 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.2 | 1.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.2 | 1.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 1.5 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 0.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 0.6 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 1.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 0.8 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.2 | 1.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 1.3 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.2 | 0.5 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 2.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 2.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.7 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.5 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.8 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 3.8 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 0.5 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.1 | 0.6 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 2.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 21.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 5.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.5 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.3 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 4.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.4 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 1.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.8 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 2.2 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.1 | 0.4 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.1 | 0.3 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.9 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.8 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.8 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.9 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 4.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.5 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.8 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.6 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 3.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.5 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 0.7 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.7 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 0.3 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 4.0 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.2 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.4 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 1.0 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.4 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 0.4 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.4 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.8 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.5 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.0 | 1.0 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.9 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 3.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 2.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 1.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 6.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 1.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 3.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 2.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 3.0 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.1 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 5.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.0 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 3.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 12.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.5 | 29.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.4 | 5.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 3.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 3.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 2.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 0.5 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.1 | 1.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.0 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 2.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.9 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.1 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |