Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
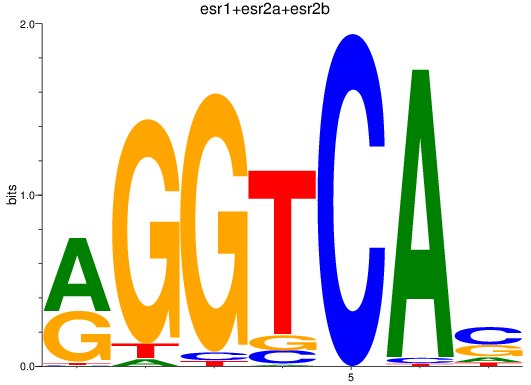
Results for esr1+esr2a+esr2b
Z-value: 0.38
Transcription factors associated with esr1+esr2a+esr2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
esr1
|
ENSDARG00000004111 | estrogen receptor 1 |
|
esr2a
|
ENSDARG00000016454 | estrogen receptor 2a |
|
esr2b
|
ENSDARG00000034181 | estrogen receptor 2b |
|
esr1
|
ENSDARG00000112357 | estrogen receptor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| esr2b | dr11_v1_chr13_+_37022601_37022632 | 0.62 | 5.8e-03 | Click! |
| esr1 | dr11_v1_chr20_-_26382284_26382284 | 0.53 | 2.3e-02 | Click! |
| esr2a | dr11_v1_chr20_+_21583639_21583639 | -0.14 | 5.7e-01 | Click! |
Activity profile of esr1+esr2a+esr2b motif
Sorted Z-values of esr1+esr2a+esr2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_44027391 | 1.03 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr12_+_49125510 | 0.95 |
ENSDART00000185804
|
FO704607.1
|
|
| chr5_-_71722257 | 0.85 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr5_+_62723233 | 0.77 |
ENSDART00000183718
|
nanos2
|
nanos homolog 2 |
| chr13_+_50375800 | 0.73 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr1_-_44704261 | 0.73 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr20_+_54738210 | 0.69 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr1_-_30039331 | 0.69 |
ENSDART00000086935
ENSDART00000143800 |
MARCH4 (1 of many)
|
zgc:153256 |
| chr4_+_17280868 | 0.69 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr12_-_47782623 | 0.67 |
ENSDART00000115742
|
selenou1b
|
selenoprotein U1b |
| chr8_+_24745041 | 0.67 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr12_+_7491690 | 0.63 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr4_-_191736 | 0.61 |
ENSDART00000169187
ENSDART00000192054 |
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr12_-_27242498 | 0.61 |
ENSDART00000152609
ENSDART00000152170 |
si:dkey-11c5.11
|
si:dkey-11c5.11 |
| chr2_-_38000276 | 0.59 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr1_-_14694809 | 0.57 |
ENSDART00000193331
|
CU138548.5
|
|
| chr7_+_29012033 | 0.51 |
ENSDART00000173909
ENSDART00000145762 |
dnaaf1
|
dynein, axonemal, assembly factor 1 |
| chr6_-_40352215 | 0.50 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr7_+_39720317 | 0.49 |
ENSDART00000164996
|
si:dkey-58i8.5
|
si:dkey-58i8.5 |
| chr3_+_28953274 | 0.49 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr5_+_42467867 | 0.48 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr23_+_42810055 | 0.48 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr23_-_39636195 | 0.47 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr9_-_48736388 | 0.47 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr11_+_5842632 | 0.47 |
ENSDART00000111374
ENSDART00000158599 |
ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, (NADH-coenzyme Q reductase) |
| chr17_+_46739693 | 0.46 |
ENSDART00000097810
|
pimr22
|
Pim proto-oncogene, serine/threonine kinase, related 22 |
| chr1_-_59176949 | 0.45 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr8_+_23213320 | 0.45 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr4_+_38344 | 0.45 |
ENSDART00000170197
ENSDART00000175348 |
phtf2
|
putative homeodomain transcription factor 2 |
| chr5_+_70155935 | 0.45 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr8_+_21406769 | 0.45 |
ENSDART00000135766
|
si:dkey-163f12.6
|
si:dkey-163f12.6 |
| chr22_-_2886937 | 0.44 |
ENSDART00000063533
|
aqp12
|
aquaporin 12 |
| chr13_-_36703164 | 0.44 |
ENSDART00000044357
|
cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr15_-_28095532 | 0.44 |
ENSDART00000191490
|
cryba1a
|
crystallin, beta A1a |
| chr17_+_42027969 | 0.44 |
ENSDART00000147563
|
kiz
|
kizuna centrosomal protein |
| chr17_-_39786222 | 0.43 |
ENSDART00000154515
|
pimr62
|
Pim proto-oncogene, serine/threonine kinase, related 62 |
| chr1_+_57311901 | 0.42 |
ENSDART00000149397
|
mycbpap
|
mycbp associated protein |
| chr5_+_20693724 | 0.42 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr16_+_10963602 | 0.41 |
ENSDART00000141032
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr24_-_39610585 | 0.41 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr25_+_4954220 | 0.41 |
ENSDART00000156034
|
si:ch73-265h17.2
|
si:ch73-265h17.2 |
| chr6_-_49873020 | 0.41 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr5_+_2815021 | 0.40 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr20_+_1398564 | 0.40 |
ENSDART00000002242
|
leg1.2
|
liver-enriched gene 1, tandem duplicate 2 |
| chr16_+_41517188 | 0.39 |
ENSDART00000049976
|
si:dkey-11p23.7
|
si:dkey-11p23.7 |
| chr7_-_51032128 | 0.39 |
ENSDART00000182781
ENSDART00000121574 |
col4a6
|
collagen, type IV, alpha 6 |
| chr1_+_54626491 | 0.39 |
ENSDART00000136063
|
si:ch211-202h22.9
|
si:ch211-202h22.9 |
| chr18_+_31016379 | 0.38 |
ENSDART00000172461
ENSDART00000163471 |
uraha
|
urate (5-hydroxyiso-) hydrolase a |
| chr4_-_5826320 | 0.38 |
ENSDART00000165354
|
foxm1
|
forkhead box M1 |
| chr14_-_215051 | 0.38 |
ENSDART00000054822
|
nkx3.2
|
NK3 homeobox 2 |
| chr23_-_45568816 | 0.38 |
ENSDART00000076086
|
cyp17a2
|
cytochrome P450, family 17, subfamily A, polypeptide 2 |
| chr6_+_12853655 | 0.37 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr20_-_43462190 | 0.37 |
ENSDART00000100697
|
pimr133
|
Pim proto-oncogene, serine/threonine kinase, related 133 |
| chr21_-_25756119 | 0.37 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr1_-_17650223 | 0.37 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr4_+_18489207 | 0.35 |
ENSDART00000135276
|
si:dkey-202b22.5
|
si:dkey-202b22.5 |
| chr23_-_45319592 | 0.35 |
ENSDART00000189861
|
ccdc171
|
coiled-coil domain containing 171 |
| chr25_+_3326885 | 0.34 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr13_+_8677166 | 0.34 |
ENSDART00000181016
ENSDART00000135028 |
prop1
|
PROP paired-like homeobox 1 |
| chr23_+_42813415 | 0.34 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr5_-_51148298 | 0.34 |
ENSDART00000042420
|
ccdc180
|
coiled-coil domain containing 180 |
| chr18_+_7456888 | 0.34 |
ENSDART00000081468
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr6_+_7250824 | 0.34 |
ENSDART00000177226
|
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr11_-_11471857 | 0.34 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr9_-_710896 | 0.34 |
ENSDART00000180478
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr12_+_18533198 | 0.33 |
ENSDART00000189729
|
meiob
|
meiosis specific with OB domains |
| chr11_+_24815927 | 0.33 |
ENSDART00000146838
|
rabif
|
RAB interacting factor |
| chr11_-_24681292 | 0.33 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr13_+_21826369 | 0.33 |
ENSDART00000165150
ENSDART00000192115 |
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr7_+_568819 | 0.32 |
ENSDART00000173716
|
nrxn2b
|
neurexin 2b |
| chr3_+_1182315 | 0.32 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr8_+_54188978 | 0.32 |
ENSDART00000175913
|
FO904970.1
|
|
| chr16_+_23972126 | 0.32 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr22_-_15593824 | 0.32 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr8_+_39760258 | 0.32 |
ENSDART00000037914
|
cox6a1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr23_-_45705525 | 0.32 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr15_-_22251089 | 0.32 |
ENSDART00000153646
|
si:ch211-286b4.4
|
si:ch211-286b4.4 |
| chr9_+_52613820 | 0.32 |
ENSDART00000168753
ENSDART00000165580 ENSDART00000164769 |
si:ch211-241j8.2
|
si:ch211-241j8.2 |
| chr11_+_6010177 | 0.32 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr6_+_475264 | 0.31 |
ENSDART00000193615
|
LO017974.1
|
|
| chr11_+_24816238 | 0.31 |
ENSDART00000115356
|
rabif
|
RAB interacting factor |
| chr8_-_49431939 | 0.30 |
ENSDART00000011453
ENSDART00000088240 ENSDART00000114173 |
sypb
|
synaptophysin b |
| chr18_-_44285539 | 0.30 |
ENSDART00000137222
|
pimr179
|
Pim proto-oncogene, serine/threonine kinase, related 179 |
| chr16_+_4838808 | 0.30 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr18_+_62932 | 0.30 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr23_-_31372639 | 0.30 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr3_-_61592417 | 0.29 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr11_-_2478374 | 0.29 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr13_-_9525527 | 0.29 |
ENSDART00000190618
|
CR848040.5
|
|
| chr8_+_46536893 | 0.29 |
ENSDART00000124023
|
pimr187
|
Pim proto-oncogene, serine/threonine kinase, related 187 |
| chr25_-_16600811 | 0.29 |
ENSDART00000011397
|
cpa2
|
carboxypeptidase A2 (pancreatic) |
| chr22_-_38274188 | 0.29 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr23_+_44634187 | 0.29 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr20_+_18232893 | 0.29 |
ENSDART00000189225
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr3_-_13147310 | 0.28 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr11_+_30162407 | 0.28 |
ENSDART00000190333
ENSDART00000127502 |
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr14_-_8724290 | 0.28 |
ENSDART00000161171
|
pimr56
|
Pim proto-oncogene, serine/threonine kinase, related 56 |
| chr5_-_1963498 | 0.28 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr23_-_38054 | 0.28 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr8_+_14158021 | 0.28 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr6_+_26346686 | 0.28 |
ENSDART00000154183
|
dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr6_+_3280939 | 0.28 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr17_-_17764801 | 0.27 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr3_+_62196672 | 0.27 |
ENSDART00000097312
|
sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr19_+_46095210 | 0.27 |
ENSDART00000159753
|
stmnd1
|
stathmin domain containing 1 |
| chr21_-_45073764 | 0.27 |
ENSDART00000181390
ENSDART00000063714 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr4_-_8152746 | 0.27 |
ENSDART00000012928
ENSDART00000177482 |
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr12_+_49135755 | 0.27 |
ENSDART00000153460
|
si:zfos-911d5.4
|
si:zfos-911d5.4 |
| chr13_-_37127970 | 0.27 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr21_-_11856143 | 0.27 |
ENSDART00000151204
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr2_-_23931536 | 0.27 |
ENSDART00000121885
|
tgfbr1a
|
transforming growth factor, beta receptor 1 a |
| chr23_+_21638258 | 0.27 |
ENSDART00000104188
|
igsf21b
|
immunoglobin superfamily, member 21b |
| chr5_-_69523816 | 0.27 |
ENSDART00000112692
|
si:ch211-157p22.10
|
si:ch211-157p22.10 |
| chr18_+_50801307 | 0.26 |
ENSDART00000174126
|
tspan4a
|
tetraspanin 4a |
| chr24_-_37003577 | 0.26 |
ENSDART00000137777
|
dnaaf3
|
dynein, axonemal, assembly factor 3 |
| chr8_-_48675775 | 0.26 |
ENSDART00000060785
|
pimr183
|
Pim proto-oncogene, serine/threonine kinase, related 183 |
| chr13_-_36418921 | 0.26 |
ENSDART00000135804
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr24_-_8777781 | 0.26 |
ENSDART00000082362
ENSDART00000177400 |
mak
|
male germ cell-associated kinase |
| chr5_+_54400971 | 0.26 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr21_-_8420830 | 0.25 |
ENSDART00000138040
|
lhx2a
|
LIM homeobox 2a |
| chr25_-_3469576 | 0.25 |
ENSDART00000186738
|
hbp1
|
HMG-box transcription factor 1 |
| chr20_-_16906623 | 0.25 |
ENSDART00000012859
ENSDART00000171628 |
psma6b
|
proteasome subunit alpha 6b |
| chr20_-_43459761 | 0.25 |
ENSDART00000137422
|
pimr133
|
Pim proto-oncogene, serine/threonine kinase, related 133 |
| chr23_+_9560991 | 0.25 |
ENSDART00000081433
ENSDART00000131594 ENSDART00000130069 ENSDART00000138601 |
adrm1
|
adhesion regulating molecule 1 |
| chr17_-_46933567 | 0.25 |
ENSDART00000157274
|
pimr25
|
Pim proto-oncogene, serine/threonine kinase, related 25 |
| chr14_+_45732081 | 0.25 |
ENSDART00000110103
|
ccdc88b
|
coiled-coil domain containing 88B |
| chr22_-_35330532 | 0.25 |
ENSDART00000172654
|
FO818743.1
|
|
| chr7_+_21331688 | 0.25 |
ENSDART00000128014
|
dnah2
|
dynein, axonemal, heavy chain 2 |
| chr25_+_13731726 | 0.25 |
ENSDART00000181065
|
ccdc135
|
coiled-coil domain containing 135 |
| chr5_-_3889047 | 0.25 |
ENSDART00000185817
|
mlxipl
|
MLX interacting protein like |
| chr2_+_33051730 | 0.24 |
ENSDART00000177902
ENSDART00000187903 |
rnf220a
|
ring finger protein 220a |
| chr25_+_3327071 | 0.24 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr5_+_13353866 | 0.24 |
ENSDART00000099611
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr22_+_18816662 | 0.24 |
ENSDART00000132476
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr19_+_3826782 | 0.24 |
ENSDART00000169222
|
oscp1a
|
organic solute carrier partner 1a |
| chr13_+_51981764 | 0.24 |
ENSDART00000160698
|
klhdc3
|
kelch domain containing 3 |
| chr18_+_7594012 | 0.24 |
ENSDART00000062150
|
zgc:77752
|
zgc:77752 |
| chr11_-_6048490 | 0.24 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr3_-_30941362 | 0.24 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chr23_+_9560797 | 0.24 |
ENSDART00000180014
|
adrm1
|
adhesion regulating molecule 1 |
| chr7_-_59256806 | 0.24 |
ENSDART00000167955
|
m1ap
|
meiosis 1 associated protein |
| chr20_-_40717900 | 0.24 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr1_-_9109699 | 0.23 |
ENSDART00000147833
|
vap
|
vascular associated protein |
| chr2_-_42960353 | 0.23 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr20_-_14924858 | 0.23 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr1_+_44767657 | 0.23 |
ENSDART00000073717
|
si:dkey-9i23.4
|
si:dkey-9i23.4 |
| chrM_+_9735 | 0.23 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr10_-_17103651 | 0.23 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr19_+_712127 | 0.23 |
ENSDART00000093281
ENSDART00000180002 ENSDART00000146050 |
fhod3a
|
formin homology 2 domain containing 3a |
| chr12_+_18532866 | 0.23 |
ENSDART00000152126
ENSDART00000152443 ENSDART00000089589 |
meiob
|
meiosis specific with OB domains |
| chr1_-_8691969 | 0.23 |
ENSDART00000152153
|
abcg2b
|
ATP-binding cassette, sub-family G (WHITE), member 2b |
| chr8_-_48675411 | 0.22 |
ENSDART00000165081
|
pimr183
|
Pim proto-oncogene, serine/threonine kinase, related 183 |
| chr5_+_483965 | 0.22 |
ENSDART00000150007
|
tek
|
TEK tyrosine kinase, endothelial |
| chr19_+_4916233 | 0.22 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr2_+_56213694 | 0.22 |
ENSDART00000162582
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr7_+_40884012 | 0.22 |
ENSDART00000149395
|
shha
|
sonic hedgehog a |
| chr7_+_22293894 | 0.22 |
ENSDART00000056790
|
tmem256
|
transmembrane protein 256 |
| chr23_+_2760573 | 0.22 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr1_+_12046544 | 0.22 |
ENSDART00000152479
|
si:dkey-58j15.11
|
si:dkey-58j15.11 |
| chr5_-_36549024 | 0.22 |
ENSDART00000097671
|
zgc:158432
|
zgc:158432 |
| chr24_-_5910811 | 0.22 |
ENSDART00000163012
|
pcolce2b
|
procollagen C-endopeptidase enhancer 2b |
| chr3_-_41292275 | 0.22 |
ENSDART00000144088
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr17_+_53156530 | 0.22 |
ENSDART00000126277
ENSDART00000156774 |
dph6
|
diphthamine biosynthesis 6 |
| chr3_-_36260102 | 0.22 |
ENSDART00000126588
|
rac3a
|
Rac family small GTPase 3a |
| chr14_+_17376940 | 0.22 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr6_+_52350443 | 0.21 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr18_+_3530769 | 0.21 |
ENSDART00000163469
|
si:ch73-338a16.3
|
si:ch73-338a16.3 |
| chr21_+_11415224 | 0.21 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr19_+_58954 | 0.21 |
ENSDART00000162379
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr5_-_3839285 | 0.21 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr6_+_10871659 | 0.21 |
ENSDART00000041858
|
mtx2
|
metaxin 2 |
| chr6_+_10450000 | 0.21 |
ENSDART00000151288
ENSDART00000187431 ENSDART00000192474 ENSDART00000188214 ENSDART00000184766 ENSDART00000190082 |
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr1_+_50976975 | 0.21 |
ENSDART00000022290
ENSDART00000140982 |
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr2_-_127945 | 0.21 |
ENSDART00000056453
|
igfbp1b
|
insulin-like growth factor binding protein 1b |
| chr17_+_32374876 | 0.21 |
ENSDART00000183851
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr12_-_37653685 | 0.21 |
ENSDART00000085121
|
sdk2b
|
sidekick cell adhesion molecule 2b |
| chr14_+_17397534 | 0.20 |
ENSDART00000180162
ENSDART00000129838 |
rnf212
|
ring finger protein 212 |
| chr22_+_10201826 | 0.20 |
ENSDART00000006513
ENSDART00000132641 |
pdhb
|
pyruvate dehydrogenase E1 beta subunit |
| chr15_-_23793641 | 0.20 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr9_-_20372977 | 0.20 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr16_-_45327616 | 0.20 |
ENSDART00000158733
|
si:dkey-33i11.1
|
si:dkey-33i11.1 |
| chr6_+_9651703 | 0.20 |
ENSDART00000122937
|
tcf23
|
transcription factor 23 |
| chr24_-_25186895 | 0.20 |
ENSDART00000081029
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr16_+_21242491 | 0.20 |
ENSDART00000145886
|
osbpl3b
|
oxysterol binding protein-like 3b |
| chr2_+_49864219 | 0.20 |
ENSDART00000187744
|
rpl37
|
ribosomal protein L37 |
| chr18_-_15932704 | 0.20 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr23_+_44633858 | 0.20 |
ENSDART00000180728
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr16_-_2558653 | 0.20 |
ENSDART00000110365
|
adcy3a
|
adenylate cyclase 3a |
| chr21_+_44112914 | 0.20 |
ENSDART00000062836
|
fgf1b
|
fibroblast growth factor 1b |
| chr16_+_4839078 | 0.19 |
ENSDART00000150111
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr5_-_72376973 | 0.19 |
ENSDART00000164969
|
CABZ01100198.1
|
|
| chr18_-_48550426 | 0.19 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr10_-_27566481 | 0.19 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr6_-_37468971 | 0.19 |
ENSDART00000126379
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr13_-_50108337 | 0.19 |
ENSDART00000133308
|
nid1a
|
nidogen 1a |
| chr12_+_30234209 | 0.19 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr20_-_4731155 | 0.19 |
ENSDART00000152155
|
papola
|
poly(A) polymerase alpha |
| chr8_-_13471916 | 0.19 |
ENSDART00000146558
|
pimr105
|
Pim proto-oncogene, serine/threonine kinase, related 105 |
| chr17_+_53425037 | 0.19 |
ENSDART00000154983
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr13_+_15354012 | 0.19 |
ENSDART00000121453
|
loxl3b
|
lysyl oxidase-like 3b |
Network of associatons between targets according to the STRING database.
First level regulatory network of esr1+esr2a+esr2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 0.5 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.4 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.1 | 1.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.9 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.3 | GO:0090220 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.3 | GO:0032757 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.3 | GO:0032369 | negative regulation of lipid transport(GO:0032369) negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 0.3 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.5 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.1 | 0.4 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.1 | 0.4 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.1 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.2 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.3 | GO:0007440 | foregut morphogenesis(GO:0007440) |
| 0.1 | 0.2 | GO:0032602 | chemokine production(GO:0032602) |
| 0.1 | 0.4 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.1 | 0.3 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 0.2 | GO:0055026 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 0.2 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.3 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0072149 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.2 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.0 | 0.2 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.2 | GO:0044003 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.2 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.2 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.2 | GO:1990544 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.2 | GO:2000316 | regulation of T-helper 17 type immune response(GO:2000316) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) regulation of T-helper 17 cell lineage commitment(GO:2000328) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.1 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.7 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.1 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) response to dexamethasone(GO:0071548) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.5 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.1 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.2 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.4 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.5 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0039535 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.1 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.0 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.7 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.2 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.1 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0010634 | positive regulation of epithelial cell migration(GO:0010634) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.7 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.2 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:0016122 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.0 | 0.3 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 1.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.0 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 4.7 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.0 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:1901381 | positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 | 0.0 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.0 | 0.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.2 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.5 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.1 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 1.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.0 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.0 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 1.3 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0098533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 0.6 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.5 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.4 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 0.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.2 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.2 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.2 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.0 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.4 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.0 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |