Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
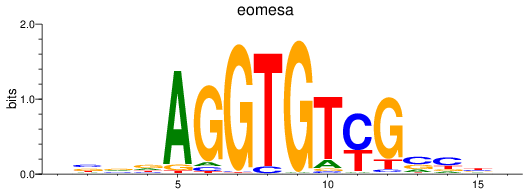
Results for eomesa
Z-value: 1.39
Transcription factors associated with eomesa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
eomesa
|
ENSDARG00000006640 | eomesodermin homolog a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| eomesa | dr11_v1_chr19_-_868187_868187 | 0.61 | 7.3e-03 | Click! |
Activity profile of eomesa motif
Sorted Z-values of eomesa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_19596214 | 6.17 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr19_-_27564458 | 5.54 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr22_+_25704430 | 4.23 |
ENSDART00000143776
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr22_+_25687525 | 4.22 |
ENSDART00000135717
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr22_+_25681911 | 4.22 |
ENSDART00000113381
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr10_+_19595009 | 4.14 |
ENSDART00000112276
|
zgc:173837
|
zgc:173837 |
| chr22_+_25693295 | 4.14 |
ENSDART00000123888
ENSDART00000150783 |
si:dkeyp-98a7.4
si:dkeyp-98a7.3
|
si:dkeyp-98a7.4 si:dkeyp-98a7.3 |
| chr22_+_25715925 | 4.10 |
ENSDART00000150650
|
si:dkeyp-98a7.7
|
si:dkeyp-98a7.7 |
| chr22_+_25753972 | 4.05 |
ENSDART00000188417
|
AL929192.2
|
|
| chr22_+_25352530 | 3.97 |
ENSDART00000176742
ENSDART00000113186 |
si:dkey-240e12.6
|
si:dkey-240e12.6 |
| chr22_+_25274712 | 3.79 |
ENSDART00000137341
|
BX950205.1
|
|
| chr10_-_25217347 | 3.46 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr19_-_867071 | 2.96 |
ENSDART00000122257
|
eomesa
|
eomesodermin homolog a |
| chr22_+_25774750 | 2.76 |
ENSDART00000174421
|
AL929192.1
|
|
| chr11_-_1550709 | 2.23 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr15_-_35112937 | 2.15 |
ENSDART00000154565
ENSDART00000099642 |
zgc:77118
|
zgc:77118 |
| chr22_+_39096911 | 2.01 |
ENSDART00000157127
ENSDART00000153841 |
lmcd1
|
LIM and cysteine-rich domains 1 |
| chr5_+_37749503 | 1.98 |
ENSDART00000148586
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr15_-_45538773 | 1.80 |
ENSDART00000113494
|
MB21D2
|
Mab-21 domain containing 2 |
| chr22_-_36690742 | 1.30 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr11_+_14312260 | 1.28 |
ENSDART00000102518
|
si:ch211-262i1.5
|
si:ch211-262i1.5 |
| chr9_-_21238616 | 1.26 |
ENSDART00000191840
ENSDART00000189127 |
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr7_+_27691647 | 1.22 |
ENSDART00000079091
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr12_-_17424162 | 1.18 |
ENSDART00000079144
ENSDART00000079138 |
ptenb
|
phosphatase and tensin homolog B |
| chr3_+_59051503 | 1.16 |
ENSDART00000160767
|
rasd4
|
rasd family member 4 |
| chr13_-_7031033 | 1.13 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr23_-_31648026 | 1.10 |
ENSDART00000133569
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr23_-_31647793 | 1.07 |
ENSDART00000145621
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr20_-_14665002 | 1.07 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr23_+_37579107 | 1.05 |
ENSDART00000169376
|
plekhg5b
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5b |
| chr9_-_23824290 | 0.99 |
ENSDART00000059209
|
wdfy2
|
WD repeat and FYVE domain containing 2 |
| chr12_-_25380028 | 0.97 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr10_-_8956973 | 0.97 |
ENSDART00000189564
|
mocs2
|
molybdenum cofactor synthesis 2 |
| chr2_-_9607879 | 0.95 |
ENSDART00000056899
|
txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr1_-_40016058 | 0.90 |
ENSDART00000165373
|
cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr7_+_17716601 | 0.88 |
ENSDART00000173792
ENSDART00000080825 |
rtn3
|
reticulon 3 |
| chr5_+_37379825 | 0.84 |
ENSDART00000171826
|
klhl13
|
kelch-like family member 13 |
| chr19_-_81851 | 0.84 |
ENSDART00000172319
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr18_-_20444296 | 0.82 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr13_-_280827 | 0.73 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr13_-_280652 | 0.72 |
ENSDART00000193627
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr5_-_67762434 | 0.68 |
ENSDART00000167301
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr21_-_14815952 | 0.67 |
ENSDART00000134278
ENSDART00000067004 |
phpt1
|
phosphohistidine phosphatase 1 |
| chr5_-_41645058 | 0.67 |
ENSDART00000051092
|
riok2
|
RIO kinase 2 (yeast) |
| chr15_-_35126632 | 0.66 |
ENSDART00000187324
ENSDART00000037389 |
zgc:77118
zgc:55413
|
zgc:77118 zgc:55413 |
| chr10_-_8956742 | 0.60 |
ENSDART00000150192
ENSDART00000149493 |
mocs2
|
molybdenum cofactor synthesis 2 |
| chr22_-_9896180 | 0.60 |
ENSDART00000138343
|
znf990
|
zinc finger protein 990 |
| chr1_-_40015782 | 0.57 |
ENSDART00000157425
ENSDART00000159238 |
cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr13_+_33703784 | 0.56 |
ENSDART00000173361
|
macrod2
|
MACRO domain containing 2 |
| chr9_-_12885201 | 0.52 |
ENSDART00000124957
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr16_+_25107344 | 0.50 |
ENSDART00000033211
|
zgc:66448
|
zgc:66448 |
| chr7_+_21275152 | 0.50 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr17_-_33416020 | 0.50 |
ENSDART00000140149
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr25_-_23583101 | 0.47 |
ENSDART00000149107
ENSDART00000103704 ENSDART00000184903 |
nap1l4a
|
nucleosome assembly protein 1-like 4a |
| chr17_+_23554932 | 0.46 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr17_-_33415740 | 0.45 |
ENSDART00000135218
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr4_+_56833071 | 0.45 |
ENSDART00000157504
|
si:dkeyp-33c10.7
|
si:dkeyp-33c10.7 |
| chr15_+_22448456 | 0.45 |
ENSDART00000040542
ENSDART00000190270 ENSDART00000185897 |
arhgef12a
|
Rho guanine nucleotide exchange factor (GEF) 12a |
| chr24_-_36196077 | 0.44 |
ENSDART00000154395
|
esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr19_+_3849378 | 0.43 |
ENSDART00000166218
ENSDART00000159228 |
oscp1a
|
organic solute carrier partner 1a |
| chr18_+_6641542 | 0.42 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr10_+_16225870 | 0.41 |
ENSDART00000164647
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr10_+_16225553 | 0.40 |
ENSDART00000129844
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr14_+_49231776 | 0.34 |
ENSDART00000169096
|
rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| chr4_+_77948517 | 0.30 |
ENSDART00000149305
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr16_+_23947196 | 0.30 |
ENSDART00000103190
ENSDART00000132961 ENSDART00000147690 ENSDART00000142168 |
apoa4b.2
|
apolipoprotein A-IV b, tandem duplicate 2 |
| chr1_-_8966046 | 0.28 |
ENSDART00000136198
|
socs1b
|
suppressor of cytokine signaling 1b |
| chr21_-_1742159 | 0.28 |
ENSDART00000151049
|
onecut2
|
one cut homeobox 2 |
| chr23_+_24957008 | 0.28 |
ENSDART00000181069
|
nol9
|
nucleolar protein 9 |
| chr4_-_77624155 | 0.28 |
ENSDART00000099761
|
si:ch211-250m6.7
|
si:ch211-250m6.7 |
| chr22_-_18544701 | 0.25 |
ENSDART00000132156
|
cirbpb
|
cold inducible RNA binding protein b |
| chr1_-_56951369 | 0.24 |
ENSDART00000152294
|
si:ch211-1f22.2
|
si:ch211-1f22.2 |
| chr1_+_29741843 | 0.24 |
ENSDART00000136066
|
raph1b
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1b |
| chr20_-_46773258 | 0.24 |
ENSDART00000060653
|
si:ch211-57i17.5
|
si:ch211-57i17.5 |
| chr16_+_4654333 | 0.24 |
ENSDART00000167665
|
LIN28A
|
si:ch1073-284b18.2 |
| chr7_+_21887787 | 0.23 |
ENSDART00000162252
|
pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr13_-_51952020 | 0.23 |
ENSDART00000181079
|
LT631676.1
|
|
| chr11_-_40504170 | 0.22 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr24_-_24060460 | 0.20 |
ENSDART00000142813
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr17_-_27048537 | 0.20 |
ENSDART00000050018
ENSDART00000193861 |
cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr15_-_1584137 | 0.20 |
ENSDART00000056772
|
trim59
|
tripartite motif containing 59 |
| chr7_+_30493684 | 0.18 |
ENSDART00000027466
|
mindy2
|
MINDY lysine 48 deubiquitinase 2 |
| chr5_+_13647288 | 0.18 |
ENSDART00000099660
ENSDART00000139199 |
h2afva
|
H2A histone family, member Va |
| chr1_+_45323142 | 0.17 |
ENSDART00000132210
|
emp1
|
epithelial membrane protein 1 |
| chr3_+_16449490 | 0.15 |
ENSDART00000141467
|
kcnj12b
|
potassium inwardly-rectifying channel, subfamily J, member 12b |
| chr7_-_61845282 | 0.13 |
ENSDART00000182586
|
HTRA3
|
HtrA serine peptidase 3 |
| chr4_+_10721795 | 0.13 |
ENSDART00000136000
ENSDART00000067253 |
stab2
|
stabilin 2 |
| chr7_-_5396154 | 0.12 |
ENSDART00000172980
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr14_-_2361692 | 0.11 |
ENSDART00000167696
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.4 |
| chr2_-_32637592 | 0.10 |
ENSDART00000136353
|
si:dkeyp-73d8.8
|
si:dkeyp-73d8.8 |
| chr22_+_19111444 | 0.10 |
ENSDART00000109655
|
fgf22
|
fibroblast growth factor 22 |
| chr16_+_44314097 | 0.09 |
ENSDART00000148684
|
dpys
|
dihydropyrimidinase |
| chr4_-_18211 | 0.09 |
ENSDART00000171737
|
ptpn12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr25_-_17367578 | 0.08 |
ENSDART00000064591
ENSDART00000110692 |
cyp2x6
|
cytochrome P450, family 2, subfamily X, polypeptide 6 |
| chr4_+_77948970 | 0.08 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr1_+_45323400 | 0.07 |
ENSDART00000148906
ENSDART00000132366 |
emp1
|
epithelial membrane protein 1 |
| chr2_-_13216269 | 0.07 |
ENSDART00000149947
|
bcl2b
|
BCL2, apoptosis regulator b |
| chr1_+_57142509 | 0.07 |
ENSDART00000146799
|
si:ch73-94k4.3
|
si:ch73-94k4.3 |
| chr24_-_24060632 | 0.07 |
ENSDART00000090514
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr1_-_57129179 | 0.06 |
ENSDART00000157226
ENSDART00000152469 |
si:ch73-94k4.2
|
si:ch73-94k4.2 |
| chr18_+_5618368 | 0.05 |
ENSDART00000159945
|
ulk3
|
unc-51 like kinase 3 |
| chr7_-_69983462 | 0.05 |
ENSDART00000123380
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr10_+_26612321 | 0.05 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr8_-_22965214 | 0.04 |
ENSDART00000148178
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr20_-_35052464 | 0.04 |
ENSDART00000037195
|
kif26bb
|
kinesin family member 26Bb |
| chr22_-_6066867 | 0.03 |
ENSDART00000142383
|
si:dkey-19a16.1
|
si:dkey-19a16.1 |
| chr18_-_50921002 | 0.02 |
ENSDART00000017053
ENSDART00000008696 |
cttn
|
cortactin |
| chr1_-_45042210 | 0.02 |
ENSDART00000073694
|
smu1b
|
SMU1, DNA replication regulator and spliceosomal factor b |
| chr19_+_2275019 | 0.01 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr11_-_13341051 | 0.01 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
Network of associatons between targets according to the STRING database.
First level regulatory network of eomesa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.3 | 3.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.2 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.2 | 0.7 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.2 | 1.2 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.2 | 0.9 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.2 | 1.6 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.8 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 1.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.4 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 0.8 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 1.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) response to copper ion(GO:0046688) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 1.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.3 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.4 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 1.1 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 1.2 | GO:0031101 | fin regeneration(GO:0031101) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 2.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.3 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.3 | 1.2 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.3 | 1.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.3 | 1.3 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 0.9 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 3.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 0.7 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 30.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.8 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 2.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 1.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 2.5 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.2 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |