Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
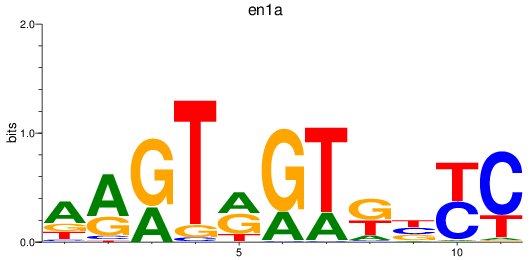
Results for en1a
Z-value: 0.30
Transcription factors associated with en1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
en1a
|
ENSDARG00000014321 | engrailed homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| en1a | dr11_v1_chr9_-_785444_785444 | 0.93 | 1.3e-08 | Click! |
Activity profile of en1a motif
Sorted Z-values of en1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_9525527 | 0.62 |
ENSDART00000190618
|
CR848040.5
|
|
| chr1_-_59287410 | 0.56 |
ENSDART00000158011
ENSDART00000170580 |
col5a3b
|
collagen, type V, alpha 3b |
| chr3_+_39922602 | 0.53 |
ENSDART00000193937
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr7_-_6470431 | 0.52 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr13_-_15700060 | 0.52 |
ENSDART00000170689
ENSDART00000010986 ENSDART00000101741 ENSDART00000139124 |
ckba
|
creatine kinase, brain a |
| chr5_-_51148298 | 0.51 |
ENSDART00000042420
|
ccdc180
|
coiled-coil domain containing 180 |
| chr15_-_45246911 | 0.49 |
ENSDART00000189557
ENSDART00000185291 |
CABZ01072607.1
|
|
| chr4_+_912563 | 0.49 |
ENSDART00000103631
|
ripply2
|
ripply transcriptional repressor 2 |
| chr5_-_23200880 | 0.48 |
ENSDART00000051531
|
iqcd
|
IQ motif containing D |
| chr2_+_22531185 | 0.43 |
ENSDART00000171959
|
hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr21_-_25618175 | 0.42 |
ENSDART00000133512
|
fosl1b
|
FOS-like antigen 1b |
| chr13_+_15215633 | 0.42 |
ENSDART00000133041
|
pank2
|
pantothenate kinase 2 |
| chr7_-_10560964 | 0.41 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr14_-_21123551 | 0.41 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr17_-_12407106 | 0.39 |
ENSDART00000183540
|
ankef1b
|
ankyrin repeat and EF-hand domain containing 1b |
| chr11_-_44876005 | 0.39 |
ENSDART00000192006
|
opn6a
|
opsin 6, group member a |
| chr2_+_33726862 | 0.39 |
ENSDART00000146745
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr7_+_44713135 | 0.38 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr17_-_6349044 | 0.38 |
ENSDART00000081707
|
oct2
|
organic cation transporter 2 |
| chr19_-_23452345 | 0.38 |
ENSDART00000136875
|
pimr82
|
Pim proto-oncogene, serine/threonine kinase, related 82 |
| chrM_+_10590 | 0.37 |
ENSDART00000093615
|
mt-nd3
|
NADH dehydrogenase 3, mitochondrial |
| chr20_-_43495506 | 0.36 |
ENSDART00000170679
|
pimr132
|
Pim proto-oncogene, serine/threonine kinase, related 132 |
| chr2_+_33751490 | 0.36 |
ENSDART00000145031
|
si:dkey-31m5.7
|
si:dkey-31m5.7 |
| chr4_+_14660769 | 0.35 |
ENSDART00000168152
ENSDART00000013990 ENSDART00000079987 |
abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr7_+_38507006 | 0.35 |
ENSDART00000173830
|
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr12_+_27096835 | 0.35 |
ENSDART00000149475
|
ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr14_+_45732081 | 0.34 |
ENSDART00000110103
|
ccdc88b
|
coiled-coil domain containing 88B |
| chr19_+_25476228 | 0.33 |
ENSDART00000112808
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr15_-_19771981 | 0.32 |
ENSDART00000175502
ENSDART00000159475 |
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr23_+_42819221 | 0.31 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr5_-_68022631 | 0.31 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr7_+_49695904 | 0.30 |
ENSDART00000183550
ENSDART00000126991 |
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr19_+_9091673 | 0.30 |
ENSDART00000052898
|
si:ch211-81a5.5
|
si:ch211-81a5.5 |
| chr8_+_26565512 | 0.30 |
ENSDART00000140980
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr20_-_52631998 | 0.29 |
ENSDART00000145283
ENSDART00000139072 |
si:ch211-221n20.1
|
si:ch211-221n20.1 |
| chr2_+_33697340 | 0.29 |
ENSDART00000137669
|
si:dkey-31m5.3
|
si:dkey-31m5.3 |
| chr5_+_66353589 | 0.29 |
ENSDART00000138246
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr13_+_27314795 | 0.28 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr21_+_25198637 | 0.28 |
ENSDART00000164972
|
si:dkey-183i3.6
|
si:dkey-183i3.6 |
| chr1_+_12009673 | 0.28 |
ENSDART00000080100
|
slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr19_-_31402429 | 0.26 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr16_+_4838808 | 0.26 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr7_+_49664174 | 0.25 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr25_-_554142 | 0.24 |
ENSDART00000028997
|
myo9ab
|
myosin IXAb |
| chr20_+_23390984 | 0.23 |
ENSDART00000136922
|
fryl
|
furry homolog, like |
| chr19_-_10648523 | 0.23 |
ENSDART00000059358
|
tekt2
|
tektin 2 (testicular) |
| chr4_-_5831522 | 0.23 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr2_-_16562505 | 0.22 |
ENSDART00000156406
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr10_+_33982010 | 0.22 |
ENSDART00000180431
|
fryb
|
furry homolog b (Drosophila) |
| chr4_-_76154252 | 0.22 |
ENSDART00000161269
|
BX957252.1
|
|
| chr4_-_52165969 | 0.22 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr8_-_50147948 | 0.22 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr22_-_37007365 | 0.22 |
ENSDART00000157966
|
ATP11B
|
si:dkey-211e20.10 |
| chr20_+_16750177 | 0.22 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr5_+_69716458 | 0.22 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr12_-_37734973 | 0.21 |
ENSDART00000140353
|
sdk2b
|
sidekick cell adhesion molecule 2b |
| chr8_-_48675411 | 0.21 |
ENSDART00000165081
|
pimr183
|
Pim proto-oncogene, serine/threonine kinase, related 183 |
| chr10_+_28428222 | 0.21 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr3_-_32902138 | 0.20 |
ENSDART00000144026
ENSDART00000083874 ENSDART00000145443 ENSDART00000148239 ENSDART00000134917 |
kat7a
|
K(lysine) acetyltransferase 7a |
| chr21_-_14803366 | 0.20 |
ENSDART00000190872
|
si:dkey-11o18.5
|
si:dkey-11o18.5 |
| chr1_+_10129099 | 0.20 |
ENSDART00000187740
|
rbm46
|
RNA binding motif protein 46 |
| chr15_-_34558579 | 0.20 |
ENSDART00000140882
|
ispd
|
isoprenoid synthase domain containing |
| chr4_+_13615731 | 0.19 |
ENSDART00000141395
ENSDART00000144310 |
ccdc87
|
coiled-coil domain containing 87 |
| chr20_+_36629173 | 0.19 |
ENSDART00000161241
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr2_-_24554416 | 0.19 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr10_-_16062565 | 0.18 |
ENSDART00000188728
|
si:dkey-184a18.5
|
si:dkey-184a18.5 |
| chr15_+_714203 | 0.18 |
ENSDART00000153847
|
si:dkey-7i4.24
|
si:dkey-7i4.24 |
| chr6_-_39764995 | 0.18 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr3_-_21166597 | 0.18 |
ENSDART00000175941
|
taok2a
|
TAO kinase 2a |
| chr3_+_3519607 | 0.18 |
ENSDART00000150922
|
cdc42ep1b
|
CDC42 effector protein (Rho GTPase binding) 1b |
| chr22_-_20733822 | 0.17 |
ENSDART00000133780
|
si:dkey-3k20.1
|
si:dkey-3k20.1 |
| chr1_-_45614481 | 0.17 |
ENSDART00000148413
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr7_+_3489195 | 0.17 |
ENSDART00000183876
|
si:ch211-285c6.3
|
si:ch211-285c6.3 |
| chr7_+_29954709 | 0.17 |
ENSDART00000173904
|
tpma
|
alpha-tropomyosin |
| chrM_-_15232 | 0.17 |
ENSDART00000093623
|
mt-nd6
|
NADH dehydrogenase 6, mitochondrial |
| chr6_+_11369135 | 0.17 |
ENSDART00000151260
|
zgc:112416
|
zgc:112416 |
| chr21_-_40676224 | 0.17 |
ENSDART00000162623
|
arxb
|
aristaless related homeobox b |
| chr20_+_539852 | 0.17 |
ENSDART00000185994
|
dse
|
dermatan sulfate epimerase |
| chr6_+_39184236 | 0.17 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr15_-_19772372 | 0.17 |
ENSDART00000152729
|
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr5_-_55625247 | 0.16 |
ENSDART00000133156
|
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr19_-_9882821 | 0.16 |
ENSDART00000147128
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr3_-_32934359 | 0.16 |
ENSDART00000124160
|
aoc2
|
amine oxidase, copper containing 2 |
| chr1_+_34295925 | 0.16 |
ENSDART00000075584
|
kctd12.2
|
potassium channel tetramerisation domain containing 12.2 |
| chr5_+_22459087 | 0.15 |
ENSDART00000134781
|
BX546499.1
|
|
| chr18_+_17624295 | 0.15 |
ENSDART00000151865
ENSDART00000190408 ENSDART00000189204 ENSDART00000188894 ENSDART00000033762 ENSDART00000193471 |
nlrc5
|
NLR family, CARD domain containing 5 |
| chrM_+_12897 | 0.15 |
ENSDART00000093622
|
mt-nd5
|
NADH dehydrogenase 5, mitochondrial |
| chr24_+_9372292 | 0.15 |
ENSDART00000082422
ENSDART00000191127 ENSDART00000180510 |
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr7_-_60096318 | 0.15 |
ENSDART00000189125
|
BX511067.1
|
|
| chr25_-_3469576 | 0.15 |
ENSDART00000186738
|
hbp1
|
HMG-box transcription factor 1 |
| chr10_+_38708099 | 0.15 |
ENSDART00000172306
|
tmprss2
|
transmembrane protease, serine 2 |
| chr16_+_26017360 | 0.15 |
ENSDART00000149466
|
prss59.2
|
protease, serine, 59, tandem duplicate 2 |
| chr1_+_33647682 | 0.14 |
ENSDART00000114384
|
znf654
|
zinc finger protein 654 |
| chr19_-_3240605 | 0.14 |
ENSDART00000105168
|
si:ch211-133n4.4
|
si:ch211-133n4.4 |
| chr12_+_34119439 | 0.14 |
ENSDART00000032821
|
cyth1b
|
cytohesin 1b |
| chr2_-_985417 | 0.14 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr19_-_18626515 | 0.14 |
ENSDART00000160624
|
rps18
|
ribosomal protein S18 |
| chr10_+_38512270 | 0.13 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr16_+_24626448 | 0.13 |
ENSDART00000153591
|
si:dkey-56f14.7
|
si:dkey-56f14.7 |
| chr13_-_45201300 | 0.13 |
ENSDART00000074750
ENSDART00000180265 |
runx3
|
runt-related transcription factor 3 |
| chr12_-_8070969 | 0.13 |
ENSDART00000020995
|
tmem26b
|
transmembrane protein 26b |
| chr6_+_46320693 | 0.13 |
ENSDART00000128621
ENSDART00000155917 |
si:dkeyp-67f1.2
|
si:dkeyp-67f1.2 |
| chr25_+_7504314 | 0.13 |
ENSDART00000163231
|
ifitm5
|
interferon induced transmembrane protein 5 |
| chr19_-_10730488 | 0.13 |
ENSDART00000126033
|
slc9a3.1
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3, tandem duplicate 1 |
| chr4_-_28958601 | 0.13 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr9_+_21277846 | 0.12 |
ENSDART00000139620
ENSDART00000110996 ENSDART00000111899 |
lats2
|
large tumor suppressor kinase 2 |
| chr5_+_61863996 | 0.12 |
ENSDART00000082879
|
sympk
|
symplekin |
| chr20_+_34845672 | 0.12 |
ENSDART00000128895
|
emilin1a
|
elastin microfibril interfacer 1a |
| chr1_-_11913170 | 0.12 |
ENSDART00000165543
|
grk4
|
G protein-coupled receptor kinase 4 |
| chr22_+_36547670 | 0.12 |
ENSDART00000125991
ENSDART00000186372 |
armc9
|
armadillo repeat containing 9 |
| chr2_+_30969029 | 0.12 |
ENSDART00000085242
|
lpin2
|
lipin 2 |
| chr25_+_6186823 | 0.12 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr20_-_20312789 | 0.12 |
ENSDART00000114779
|
si:ch211-212g7.6
|
si:ch211-212g7.6 |
| chr2_-_22492289 | 0.12 |
ENSDART00000168022
|
gtf2b
|
general transcription factor IIB |
| chr6_+_41446541 | 0.12 |
ENSDART00000029553
ENSDART00000128756 ENSDART00000144864 |
rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr7_-_69636502 | 0.12 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr6_+_21684296 | 0.11 |
ENSDART00000057223
|
rhebl1
|
Ras homolog, mTORC1 binding like 1 |
| chr23_+_36653376 | 0.11 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr11_+_24716837 | 0.11 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr3_-_4579214 | 0.11 |
ENSDART00000164456
|
ftr50
|
finTRIM family, member 50 |
| chr1_-_46632948 | 0.11 |
ENSDART00000148893
ENSDART00000053232 |
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr5_+_21225017 | 0.11 |
ENSDART00000141573
|
si:dkey-13n15.11
|
si:dkey-13n15.11 |
| chr5_+_58687541 | 0.11 |
ENSDART00000083015
ENSDART00000181902 |
ccdc84
|
coiled-coil domain containing 84 |
| chr2_+_45479841 | 0.10 |
ENSDART00000151856
|
si:ch211-66k16.28
|
si:ch211-66k16.28 |
| chr20_-_47027379 | 0.10 |
ENSDART00000100320
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr8_+_17168114 | 0.10 |
ENSDART00000183901
|
cenph
|
centromere protein H |
| chr16_+_36768674 | 0.10 |
ENSDART00000169208
ENSDART00000180470 |
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr1_+_32013688 | 0.10 |
ENSDART00000168045
|
BX901898.2
|
|
| chr8_+_17167876 | 0.10 |
ENSDART00000134665
|
cenph
|
centromere protein H |
| chr8_+_2456854 | 0.10 |
ENSDART00000133938
ENSDART00000002764 |
polb
|
polymerase (DNA directed), beta |
| chr3_-_4557653 | 0.10 |
ENSDART00000111305
|
ftr50
|
finTRIM family, member 50 |
| chr19_+_1370504 | 0.09 |
ENSDART00000158946
|
dgat1a
|
diacylglycerol O-acyltransferase 1a |
| chr7_-_52153105 | 0.09 |
ENSDART00000174378
|
CT737190.1
|
|
| chr12_-_24904944 | 0.09 |
ENSDART00000153436
|
fbxo11a
|
F-box protein 11a |
| chr14_+_38845674 | 0.09 |
ENSDART00000129958
|
hnrnpa0b
|
heterogeneous nuclear ribonucleoprotein A0b |
| chr19_-_10810006 | 0.09 |
ENSDART00000151157
|
si:dkey-3n22.9
|
si:dkey-3n22.9 |
| chr7_+_31879649 | 0.09 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr16_+_25809235 | 0.09 |
ENSDART00000181196
|
irgq1
|
immunity-related GTPase family, q1 |
| chr21_+_19517492 | 0.09 |
ENSDART00000123168
ENSDART00000187993 |
gzmk
|
granzyme K |
| chr25_+_28825657 | 0.09 |
ENSDART00000153625
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr15_-_20233105 | 0.09 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr22_+_19247255 | 0.09 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr7_+_31879986 | 0.09 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr16_+_41570653 | 0.09 |
ENSDART00000102665
|
aste1a
|
asteroid homolog 1a |
| chr3_+_52737565 | 0.09 |
ENSDART00000108639
|
gmip
|
GEM interacting protein |
| chr7_-_26462831 | 0.08 |
ENSDART00000113543
|
mblac1
|
metallo-beta-lactamase domain containing 1 |
| chr22_+_14051245 | 0.08 |
ENSDART00000043711
ENSDART00000164259 |
aox6
|
aldehyde oxidase 6 |
| chr5_-_28038623 | 0.08 |
ENSDART00000160013
|
zgc:113436
|
zgc:113436 |
| chr22_+_30112390 | 0.08 |
ENSDART00000191715
|
add3a
|
adducin 3 (gamma) a |
| chr2_-_51794472 | 0.08 |
ENSDART00000186652
|
BX908782.3
|
|
| chr9_+_13733468 | 0.08 |
ENSDART00000165954
ENSDART00000160355 ENSDART00000081040 ENSDART00000138254 ENSDART00000081015 ENSDART00000141314 |
abi2a
|
abl-interactor 2a |
| chr23_-_2448234 | 0.08 |
ENSDART00000082097
|
LO018513.1
|
|
| chr7_+_30178453 | 0.08 |
ENSDART00000174420
|
si:ch211-107o10.3
|
si:ch211-107o10.3 |
| chr21_+_38638979 | 0.08 |
ENSDART00000143373
|
rbmx2
|
RNA binding motif protein, X-linked 2 |
| chr18_+_41232719 | 0.08 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr12_-_10365875 | 0.08 |
ENSDART00000142386
ENSDART00000007335 |
ndc80
|
NDC80 kinetochore complex component |
| chr10_+_41765944 | 0.08 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr3_+_18046776 | 0.08 |
ENSDART00000154770
|
mettl26
|
methyltransferase like 26 |
| chr18_+_26829362 | 0.08 |
ENSDART00000132728
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr14_-_31854830 | 0.08 |
ENSDART00000148550
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr8_-_44015210 | 0.08 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr20_+_18325556 | 0.08 |
ENSDART00000123559
|
znf521
|
zinc finger protein 521 |
| chr7_-_35409027 | 0.08 |
ENSDART00000128334
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr14_-_38872536 | 0.08 |
ENSDART00000184315
ENSDART00000127479 |
gsr
|
glutathione reductase |
| chr2_+_45441563 | 0.08 |
ENSDART00000151882
|
si:ch211-66k16.27
|
si:ch211-66k16.27 |
| chr6_-_40195510 | 0.07 |
ENSDART00000156156
|
col7a1
|
collagen, type VII, alpha 1 |
| chr17_-_45734459 | 0.07 |
ENSDART00000137807
|
arf6b
|
ADP-ribosylation factor 6b |
| chr6_-_43780751 | 0.07 |
ENSDART00000188257
|
foxp1b
|
forkhead box P1b |
| chr9_+_1313418 | 0.07 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr24_-_25184553 | 0.07 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr5_-_64203101 | 0.07 |
ENSDART00000029364
|
ak5l
|
adenylate kinase 5, like |
| chr21_+_10782013 | 0.07 |
ENSDART00000146248
|
znf532
|
zinc finger protein 532 |
| chr4_-_62714083 | 0.07 |
ENSDART00000188299
|
si:dkey-28k24.2
|
si:dkey-28k24.2 |
| chr18_-_17513426 | 0.07 |
ENSDART00000146725
|
b3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr19_+_14921000 | 0.07 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr18_+_36625490 | 0.06 |
ENSDART00000148032
|
clasrp
|
CLK4-associating serine/arginine rich protein |
| chr17_-_18797245 | 0.06 |
ENSDART00000045991
|
vrk1
|
vaccinia related kinase 1 |
| chr20_-_39391833 | 0.06 |
ENSDART00000135149
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr3_+_62205858 | 0.06 |
ENSDART00000126807
|
zgc:173575
|
zgc:173575 |
| chr17_-_45734231 | 0.06 |
ENSDART00000074873
|
arf6b
|
ADP-ribosylation factor 6b |
| chr5_-_8164439 | 0.06 |
ENSDART00000189912
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr8_+_40284973 | 0.06 |
ENSDART00000018401
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr16_+_5196226 | 0.06 |
ENSDART00000189704
|
soga3a
|
SOGA family member 3a |
| chr15_+_23799461 | 0.06 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr4_+_12333168 | 0.06 |
ENSDART00000172455
ENSDART00000146166 |
pimr214
pimr171
|
Pim proto-oncogene, serine/threonine kinase, related 214 Pim proto-oncogene, serine/threonine kinase, related 171 |
| chr10_+_33744098 | 0.06 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr14_+_8476769 | 0.06 |
ENSDART00000131773
|
dnajc4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr22_+_17433968 | 0.06 |
ENSDART00000139523
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr7_-_34062301 | 0.05 |
ENSDART00000052404
|
map2k5
|
mitogen-activated protein kinase kinase 5 |
| chr18_-_19095141 | 0.05 |
ENSDART00000090962
ENSDART00000090922 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr20_+_10723292 | 0.05 |
ENSDART00000152805
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr2_+_32780138 | 0.05 |
ENSDART00000082250
|
zgc:136930
|
zgc:136930 |
| chr22_+_2183110 | 0.05 |
ENSDART00000159279
ENSDART00000121703 |
znf1152
|
zinc finger protein 1152 |
| chr21_+_10866421 | 0.05 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr5_-_30615901 | 0.05 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr3_+_26345732 | 0.05 |
ENSDART00000128613
|
rps15a
|
ribosomal protein S15a |
| chr18_-_12858016 | 0.05 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr19_+_4076259 | 0.05 |
ENSDART00000160633
|
zgc:173581
|
zgc:173581 |
| chr2_-_1514001 | 0.05 |
ENSDART00000057736
|
c8b
|
complement component 8, beta polypeptide |
| chr22_+_22303053 | 0.05 |
ENSDART00000129975
|
scamp4
|
secretory carrier membrane protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of en1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 0.4 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.2 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.1 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.5 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.2 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0044003 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.0 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.4 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.2 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.3 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:1902914 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.0 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.2 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.0 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0046620 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.5 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.4 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.2 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.5 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.3 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0022821 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.0 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |