Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
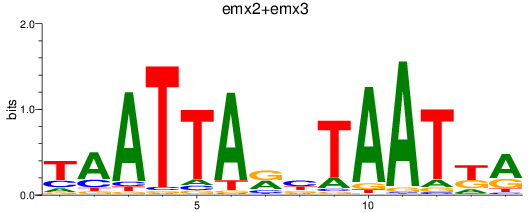
Results for emx2+emx3
Z-value: 1.19
Transcription factors associated with emx2+emx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
emx3
|
ENSDARG00000020417 | empty spiracles homeobox 3 |
|
emx2
|
ENSDARG00000039701 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| emx3 | dr11_v1_chr14_-_26377044_26377044 | -0.93 | 3.3e-08 | Click! |
| emx2 | dr11_v1_chr13_+_19322686_19322686 | -0.74 | 4.7e-04 | Click! |
Activity profile of emx2+emx3 motif
Sorted Z-values of emx2+emx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_6452444 | 5.59 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr25_+_22320738 | 4.10 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr8_-_23780334 | 3.85 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr9_+_54039006 | 3.71 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr1_-_18811517 | 3.60 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr23_+_36460239 | 3.43 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr20_+_54299419 | 3.35 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr20_+_54309148 | 3.31 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr17_-_4245902 | 3.19 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr4_+_9467049 | 3.16 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr20_+_54304800 | 2.76 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr18_-_40708537 | 2.75 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr10_-_34916208 | 2.47 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr19_-_10330778 | 2.37 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr19_-_25149034 | 2.07 |
ENSDART00000148432
ENSDART00000175266 |
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr3_+_7808459 | 2.03 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr21_+_13387965 | 2.00 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr5_+_37903790 | 1.93 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr16_-_41646164 | 1.92 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr19_-_25149598 | 1.86 |
ENSDART00000162917
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr8_-_39884359 | 1.86 |
ENSDART00000131372
|
mlec
|
malectin |
| chr19_+_14351560 | 1.85 |
ENSDART00000182732
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr14_-_25935167 | 1.83 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr10_+_26990095 | 1.75 |
ENSDART00000064111
|
faub
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed b |
| chr21_+_21743599 | 1.70 |
ENSDART00000101700
|
pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr21_-_4849029 | 1.68 |
ENSDART00000168930
ENSDART00000151019 |
notch1a
|
notch 1a |
| chr10_-_44560165 | 1.62 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr15_+_34934568 | 1.55 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr11_-_44979281 | 1.55 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr23_-_14766902 | 1.52 |
ENSDART00000168113
|
gss
|
glutathione synthetase |
| chr12_-_3978306 | 1.50 |
ENSDART00000149473
ENSDART00000114857 |
ppp4cb
|
protein phosphatase 4, catalytic subunit b |
| chr6_-_46742455 | 1.50 |
ENSDART00000011970
|
zgc:66479
|
zgc:66479 |
| chr13_+_30035253 | 1.49 |
ENSDART00000181303
ENSDART00000057525 ENSDART00000136622 |
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr15_+_34946779 | 1.48 |
ENSDART00000192661
ENSDART00000188800 ENSDART00000156515 |
si:ch73-95l15.5
zgc:55621
|
si:ch73-95l15.5 zgc:55621 |
| chr22_-_22337382 | 1.46 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr9_+_24065855 | 1.44 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr12_-_35386910 | 1.44 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr2_+_42871831 | 1.41 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr6_+_7533601 | 1.40 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr12_-_48188928 | 1.39 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr1_+_31658011 | 1.38 |
ENSDART00000192203
|
poll
|
polymerase (DNA directed), lambda |
| chr12_-_314899 | 1.34 |
ENSDART00000066579
|
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr8_+_42917515 | 1.34 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr4_-_5108844 | 1.31 |
ENSDART00000132666
ENSDART00000136096 |
tmem209
|
transmembrane protein 209 |
| chr13_-_46687097 | 1.30 |
ENSDART00000169106
ENSDART00000158202 |
CABZ01078449.1
|
|
| chr14_-_1200854 | 1.30 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr2_+_15100742 | 1.29 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr3_+_32416948 | 1.28 |
ENSDART00000157324
ENSDART00000154267 ENSDART00000186094 ENSDART00000155860 ENSDART00000156986 |
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr16_+_25068576 | 1.28 |
ENSDART00000125838
|
im:7147486
|
im:7147486 |
| chr10_-_28380919 | 1.28 |
ENSDART00000183409
ENSDART00000183105 ENSDART00000100207 ENSDART00000185392 ENSDART00000131220 |
btg3
|
B-cell translocation gene 3 |
| chr21_-_32060993 | 1.27 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr3_+_23029934 | 1.26 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr8_-_2616326 | 1.25 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr2_+_30182431 | 1.24 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr10_+_5268054 | 1.24 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr23_+_26026383 | 1.23 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr24_+_26329018 | 1.23 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr21_-_44081540 | 1.22 |
ENSDART00000130833
|
FO704810.1
|
|
| chr11_-_45171139 | 1.21 |
ENSDART00000167036
ENSDART00000161712 ENSDART00000158156 |
syngr2b
|
synaptogyrin 2b |
| chr3_+_40164129 | 1.21 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr3_+_23029484 | 1.20 |
ENSDART00000187900
|
nags
|
N-acetylglutamate synthase |
| chr21_-_2185600 | 1.20 |
ENSDART00000169897
|
zgc:171220
|
zgc:171220 |
| chr23_+_12160900 | 1.19 |
ENSDART00000136046
|
ppp1r3da
|
protein phosphatase 1, regulatory subunit 3Da |
| chr11_-_10456387 | 1.19 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr3_+_26342768 | 1.18 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr14_-_33858214 | 1.18 |
ENSDART00000112268
|
ocrl
|
oculocerebrorenal syndrome of Lowe |
| chr9_-_32300783 | 1.15 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr9_-_32300611 | 1.13 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr11_-_10456553 | 1.13 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr24_+_26328787 | 1.12 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr7_+_69459759 | 1.12 |
ENSDART00000160500
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr16_-_7793457 | 1.12 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr13_-_15082024 | 1.12 |
ENSDART00000157482
|
sfxn5a
|
sideroflexin 5a |
| chr23_-_40536017 | 1.11 |
ENSDART00000153751
ENSDART00000140623 ENSDART00000133356 |
rnf146
|
ring finger protein 146 |
| chr22_+_2751887 | 1.10 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr9_-_5318873 | 1.10 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr2_-_17392799 | 1.08 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr19_-_42045372 | 1.07 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr13_-_35761266 | 1.06 |
ENSDART00000190217
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr7_+_59033902 | 1.06 |
ENSDART00000168888
|
fastkd3
|
FAST kinase domains 3 |
| chr10_+_45089820 | 1.05 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr20_+_32552912 | 1.04 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr7_+_22313533 | 1.03 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr9_-_41040492 | 1.03 |
ENSDART00000163164
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr11_+_45110865 | 1.03 |
ENSDART00000158188
|
mgea5l
|
meningioma expressed antigen 5 (hyaluronidase) like |
| chr16_+_32736588 | 1.03 |
ENSDART00000075191
ENSDART00000168358 |
zgc:172323
|
zgc:172323 |
| chr12_+_45238292 | 1.03 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr18_+_22416051 | 1.01 |
ENSDART00000115388
|
cfdp1
|
craniofacial development protein 1 |
| chr15_+_34592215 | 0.99 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr25_-_35101673 | 0.99 |
ENSDART00000140864
|
zgc:162611
|
zgc:162611 |
| chr17_+_6469419 | 0.99 |
ENSDART00000191729
|
COQ8A (1 of many)
|
si:dkey-36g24.3 |
| chr2_-_9989919 | 0.99 |
ENSDART00000180213
ENSDART00000184369 |
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr5_-_25733745 | 0.98 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr13_+_7665890 | 0.98 |
ENSDART00000046792
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr5_-_9824908 | 0.97 |
ENSDART00000169698
|
zgc:158343
|
zgc:158343 |
| chr23_-_25135046 | 0.97 |
ENSDART00000184844
ENSDART00000103989 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr25_+_2263857 | 0.97 |
ENSDART00000076439
|
yars2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr5_-_52010122 | 0.95 |
ENSDART00000073627
ENSDART00000163898 ENSDART00000051003 |
cdk7
|
cyclin-dependent kinase 7 |
| chr20_-_38801981 | 0.94 |
ENSDART00000125333
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr9_+_38314466 | 0.92 |
ENSDART00000048753
|
ddx18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr7_-_23894879 | 0.91 |
ENSDART00000123203
|
vbp1
|
von Hippel-Lindau binding protein 1 |
| chr1_+_54124209 | 0.91 |
ENSDART00000187730
|
LO017722.1
|
|
| chr13_+_33268657 | 0.90 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr14_-_33521071 | 0.90 |
ENSDART00000052789
|
c1galt1c1
|
C1GALT1-specific chaperone 1 |
| chr25_+_20694177 | 0.89 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr1_+_45002971 | 0.88 |
ENSDART00000021336
|
dnaja1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr12_-_45238759 | 0.88 |
ENSDART00000154859
|
trim65
|
tripartite motif containing 65 |
| chr11_-_6877973 | 0.88 |
ENSDART00000160271
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr12_+_1609563 | 0.87 |
ENSDART00000163559
|
SLC39A11
|
solute carrier family 39 member 11 |
| chr16_+_24733741 | 0.87 |
ENSDART00000155217
|
si:dkey-79d12.4
|
si:dkey-79d12.4 |
| chr21_+_21195487 | 0.86 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr21_-_38618540 | 0.85 |
ENSDART00000036600
|
slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr16_-_10223741 | 0.83 |
ENSDART00000188099
|
si:rp71-15i12.1
|
si:rp71-15i12.1 |
| chr22_+_2769236 | 0.83 |
ENSDART00000141836
|
si:dkey-20i20.10
|
si:dkey-20i20.10 |
| chr2_-_31833347 | 0.83 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr3_-_15496551 | 0.82 |
ENSDART00000124063
ENSDART00000007726 |
sgf29
|
SAGA complex associated factor 29 |
| chr20_+_28803977 | 0.82 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr19_-_2822372 | 0.81 |
ENSDART00000109130
ENSDART00000122385 |
recql4
|
RecQ helicase-like 4 |
| chr6_-_39005133 | 0.80 |
ENSDART00000104116
ENSDART00000151750 |
vdrb
|
vitamin D receptor b |
| chr23_+_29966466 | 0.79 |
ENSDART00000143583
|
dvl1a
|
dishevelled segment polarity protein 1a |
| chr21_-_4250682 | 0.79 |
ENSDART00000099389
|
dnlz
|
DNL-type zinc finger |
| chr2_-_37743834 | 0.78 |
ENSDART00000088040
ENSDART00000191057 |
myo9b
|
myosin IXb |
| chr4_+_2482046 | 0.78 |
ENSDART00000103371
|
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr16_-_36064143 | 0.77 |
ENSDART00000158358
ENSDART00000182584 |
stk40
|
serine/threonine kinase 40 |
| chr21_+_6394929 | 0.77 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr24_+_19415124 | 0.77 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr2_-_17393216 | 0.77 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr1_+_6225493 | 0.77 |
ENSDART00000145378
|
fastkd2
|
FAST kinase domains 2 |
| chr6_-_7686594 | 0.76 |
ENSDART00000091836
ENSDART00000151697 |
ubn2a
|
ubinuclein 2a |
| chr2_-_40890264 | 0.76 |
ENSDART00000123886
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr11_+_31323746 | 0.76 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr2_-_40890004 | 0.76 |
ENSDART00000191746
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr2_+_1988036 | 0.76 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr20_+_26943072 | 0.76 |
ENSDART00000153215
|
cdca4
|
cell division cycle associated 4 |
| chr25_-_27621268 | 0.75 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr9_+_29643036 | 0.75 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr16_+_54209504 | 0.74 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr24_+_42004640 | 0.74 |
ENSDART00000171380
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr9_-_7390388 | 0.74 |
ENSDART00000132392
|
slc23a3
|
solute carrier family 23, member 3 |
| chr11_-_18020258 | 0.74 |
ENSDART00000156116
|
qrich1
|
glutamine-rich 1 |
| chr4_-_5019113 | 0.73 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr10_-_35257458 | 0.73 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr1_-_513762 | 0.72 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr18_+_16750080 | 0.72 |
ENSDART00000136320
|
rnf141
|
ring finger protein 141 |
| chr13_-_31008275 | 0.71 |
ENSDART00000139394
|
wdfy4
|
WDFY family member 4 |
| chr11_+_13642157 | 0.71 |
ENSDART00000060251
|
wdr18
|
WD repeat domain 18 |
| chr23_+_32101202 | 0.71 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr21_+_31253048 | 0.71 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr18_+_8833251 | 0.70 |
ENSDART00000143519
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr7_-_50410524 | 0.70 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr15_-_17813680 | 0.70 |
ENSDART00000158556
|
CT573342.2
|
|
| chr19_+_15485287 | 0.69 |
ENSDART00000182797
|
pdik1l
|
PDLIM1 interacting kinase 1 like |
| chr20_+_22799641 | 0.68 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr11_+_42474694 | 0.68 |
ENSDART00000056048
ENSDART00000184710 |
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr1_-_25144439 | 0.68 |
ENSDART00000132355
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr1_+_45663727 | 0.68 |
ENSDART00000038574
ENSDART00000141144 ENSDART00000149565 |
trappc5
|
trafficking protein particle complex 5 |
| chr6_+_30533504 | 0.67 |
ENSDART00000155842
|
wwc3
|
WWC family member 3 |
| chr7_+_15659280 | 0.67 |
ENSDART00000173414
|
mef2ab
|
myocyte enhancer factor 2ab |
| chr20_+_474288 | 0.67 |
ENSDART00000026794
|
nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr14_+_29941266 | 0.66 |
ENSDART00000112757
|
fam149a
|
family with sequence similarity 149 member A |
| chr21_+_43702016 | 0.66 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr18_+_45958375 | 0.66 |
ENSDART00000108636
|
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr25_+_3217419 | 0.65 |
ENSDART00000104859
|
rccd1
|
RCC1 domain containing 1 |
| chr25_-_25058508 | 0.65 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr12_-_3773869 | 0.65 |
ENSDART00000092983
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr16_-_30869732 | 0.64 |
ENSDART00000042716
ENSDART00000190168 |
ptk2ab
|
protein tyrosine kinase 2ab |
| chr12_+_1139690 | 0.64 |
ENSDART00000160442
|
CABZ01072885.1
|
|
| chr14_-_26425416 | 0.64 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr19_+_2685779 | 0.62 |
ENSDART00000160533
ENSDART00000097531 |
tomm7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr6_-_39218609 | 0.61 |
ENSDART00000133305
|
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr8_-_43835603 | 0.61 |
ENSDART00000134073
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr9_-_21825913 | 0.61 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr17_+_49281597 | 0.61 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr5_-_11809710 | 0.60 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr11_-_34478225 | 0.60 |
ENSDART00000189604
|
xxylt1
|
xyloside xylosyltransferase 1 |
| chr11_+_17984354 | 0.60 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr22_-_20812822 | 0.60 |
ENSDART00000193778
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr7_+_36898850 | 0.59 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr25_-_6011034 | 0.59 |
ENSDART00000075197
ENSDART00000136054 |
snx22
|
sorting nexin 22 |
| chr3_+_32118670 | 0.58 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr21_-_25395223 | 0.57 |
ENSDART00000016219
|
ppme1
|
protein phosphatase methylesterase 1 |
| chr12_+_1492641 | 0.57 |
ENSDART00000152411
|
usp22
|
ubiquitin specific peptidase 22 |
| chr17_+_8799661 | 0.56 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr7_+_66884291 | 0.55 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr18_-_2222128 | 0.55 |
ENSDART00000171402
|
pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr11_-_38533505 | 0.54 |
ENSDART00000113894
|
slc45a3
|
solute carrier family 45, member 3 |
| chr9_+_48761455 | 0.53 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr1_+_27977297 | 0.53 |
ENSDART00000180692
ENSDART00000166819 |
sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr21_-_31210749 | 0.53 |
ENSDART00000185356
|
zgc:152891
|
zgc:152891 |
| chr13_+_6219551 | 0.53 |
ENSDART00000025910
|
apmap
|
adipocyte plasma membrane associated protein |
| chr23_+_42254960 | 0.52 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr4_+_5255041 | 0.52 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr2_-_37632896 | 0.52 |
ENSDART00000008302
|
insra
|
insulin receptor a |
| chr8_+_53080515 | 0.52 |
ENSDART00000143009
|
wdr46
|
WD repeat domain 46 |
| chr7_+_36898622 | 0.52 |
ENSDART00000190773
|
tox3
|
TOX high mobility group box family member 3 |
| chr19_+_20274491 | 0.51 |
ENSDART00000090860
ENSDART00000151693 |
oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr3_+_54553931 | 0.50 |
ENSDART00000029387
|
ppan
|
peter pan homolog (Drosophila) |
| chr23_+_25135858 | 0.49 |
ENSDART00000103986
|
fam3a
|
family with sequence similarity 3, member A |
| chr23_-_900795 | 0.48 |
ENSDART00000190517
ENSDART00000182849 ENSDART00000111456 ENSDART00000185430 |
rbm10
|
RNA binding motif protein 10 |
| chr22_-_11954861 | 0.48 |
ENSDART00000131611
|
si:dkeyp-4h4.1
|
si:dkeyp-4h4.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of emx2+emx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.8 | 4.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.8 | 3.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.6 | 2.3 | GO:0045041 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.6 | 1.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.6 | 1.7 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.5 | 2.6 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.4 | 1.3 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.4 | 1.9 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.4 | 1.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 0.9 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.3 | 1.2 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.3 | 1.5 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 1.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.8 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.2 | 0.7 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.2 | 0.9 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 1.1 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.2 | 1.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.2 | 0.7 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 0.8 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 0.6 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.2 | 0.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.2 | 2.3 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.2 | 0.8 | GO:0015744 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 0.6 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 0.7 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.4 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 1.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 3.0 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 0.8 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.0 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.8 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.3 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 1.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.4 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 1.6 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 1.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.6 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.8 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.4 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 1.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.3 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.1 | 0.4 | GO:0031650 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.3 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.1 | 1.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.3 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 1.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.3 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.1 | 0.3 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.1 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.1 | 0.7 | GO:0090329 | regulation of DNA-dependent DNA replication(GO:0090329) |
| 0.1 | 0.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.2 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.1 | 0.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 1.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 2.0 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.6 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.4 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.7 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 1.6 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 1.2 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 0.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 1.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.9 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 2.5 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 0.3 | GO:0006921 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.5 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 3.7 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.8 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.6 | GO:0045005 | replication fork processing(GO:0031297) DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.0 | 0.1 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.3 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 2.0 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0042421 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 1.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.1 | GO:1902745 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.1 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.6 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.4 | GO:0098781 | ncRNA transcription(GO:0098781) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.1 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.2 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 1.3 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 1.3 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.1 | GO:2001287 | optomotor response(GO:0071632) caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 1.3 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.1 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.8 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.4 | 2.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 1.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.3 | 1.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 1.8 | GO:0035060 | brahma complex(GO:0035060) |
| 0.2 | 0.7 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.2 | 1.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 0.8 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 0.7 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 1.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.5 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.0 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 6.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 5.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.0 | GO:0098753 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 3.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.9 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.1 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.8 | 2.5 | GO:0034618 | arginine binding(GO:0034618) |
| 0.4 | 1.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.3 | 1.0 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.3 | 1.9 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.3 | 0.9 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.3 | 1.5 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 1.8 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 0.9 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.2 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 0.8 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 1.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 1.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.2 | 0.7 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 0.7 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 0.7 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 0.5 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.2 | 0.8 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.2 | 0.8 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 0.8 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.2 | 0.5 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.6 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 3.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.7 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.5 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.4 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 1.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.5 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.4 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 3.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.6 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 1.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.1 | 0.2 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.1 | 1.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.5 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.1 | 1.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.5 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 1.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.2 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 2.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 3.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 1.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 1.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 2.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.0 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 3.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 1.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.9 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 1.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 2.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.2 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 2.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 2.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.0 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 2.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 3.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 3.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 3.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 1.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 3.4 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.2 | 2.0 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 5.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.7 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.3 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |