Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for elf3
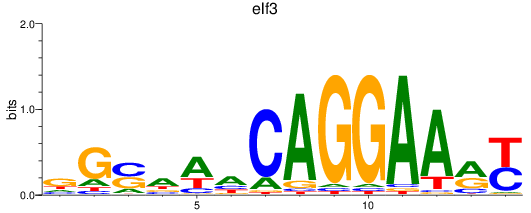
Z-value: 0.45
Transcription factors associated with elf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
elf3
|
ENSDARG00000077982 | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| elf3 | dr11_v1_chr22_+_661711_661711 | 0.59 | 9.4e-03 | Click! |
Activity profile of elf3 motif
Sorted Z-values of elf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_28727383 | 0.96 |
ENSDART00000149753
|
dcst2
|
DC-STAMP domain containing 2 |
| chr13_-_23074454 | 0.90 |
ENSDART00000065652
|
srgn
|
serglycin |
| chr5_+_42467867 | 0.84 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr20_-_43572763 | 0.78 |
ENSDART00000153251
|
pimr125
|
Pim proto-oncogene, serine/threonine kinase, related 125 |
| chr1_+_40237276 | 0.65 |
ENSDART00000037553
|
faah2a
|
fatty acid amide hydrolase 2a |
| chr9_-_48397702 | 0.63 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr13_+_33304187 | 0.63 |
ENSDART00000075826
ENSDART00000145295 |
dcdc2b
|
doublecortin domain containing 2B |
| chr17_-_10309766 | 0.61 |
ENSDART00000160994
|
ttc6
|
tetratricopeptide repeat domain 6 |
| chr3_-_32902138 | 0.54 |
ENSDART00000144026
ENSDART00000083874 ENSDART00000145443 ENSDART00000148239 ENSDART00000134917 |
kat7a
|
K(lysine) acetyltransferase 7a |
| chr1_-_59252973 | 0.54 |
ENSDART00000167061
|
si:ch1073-286c18.5
|
si:ch1073-286c18.5 |
| chr22_-_36934040 | 0.53 |
ENSDART00000151666
|
pimr206
|
Pim proto-oncogene, serine/threonine kinase, related 206 |
| chr5_-_21524251 | 0.48 |
ENSDART00000176793
ENSDART00000040184 |
tenm1
|
teneurin transmembrane protein 1 |
| chr6_-_55254786 | 0.48 |
ENSDART00000113805
|
nfatc2b
|
nuclear factor of activated T cells 2b |
| chr20_+_42761881 | 0.47 |
ENSDART00000113625
|
pimr113
|
Pim proto-oncogene, serine/threonine kinase, related 113 |
| chr5_+_42402536 | 0.47 |
ENSDART00000186754
|
BX548073.11
|
|
| chr6_-_17849786 | 0.47 |
ENSDART00000172709
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr23_-_10175898 | 0.46 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr3_+_49576662 | 0.46 |
ENSDART00000162275
|
CR394535.1
|
|
| chr20_+_39457598 | 0.46 |
ENSDART00000140931
ENSDART00000156176 |
pimr128
|
Pim proto-oncogene, serine/threonine kinase, related 128 |
| chr2_+_9061885 | 0.45 |
ENSDART00000028906
|
pigk
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr7_-_22981796 | 0.44 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr11_-_38899816 | 0.43 |
ENSDART00000141515
|
pimr127
|
Pim proto-oncogene, serine/threonine kinase, related 127 |
| chr12_-_28910419 | 0.43 |
ENSDART00000153278
ENSDART00000152937 |
CCDC189
|
si:ch73-81k8.2 |
| chr17_-_12407106 | 0.42 |
ENSDART00000183540
|
ankef1b
|
ankyrin repeat and EF-hand domain containing 1b |
| chr11_+_11842724 | 0.42 |
ENSDART00000163752
|
pimr94
|
Pim proto-oncogene, serine/threonine kinase, related 94 |
| chr20_+_34770197 | 0.41 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr22_+_12798569 | 0.40 |
ENSDART00000005720
|
stat1a
|
signal transducer and activator of transcription 1a |
| chr20_+_33739059 | 0.40 |
ENSDART00000140361
|
ppil6
|
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr20_-_43495506 | 0.40 |
ENSDART00000170679
|
pimr132
|
Pim proto-oncogene, serine/threonine kinase, related 132 |
| chr5_-_57652168 | 0.40 |
ENSDART00000166245
|
gb:eh456644
|
expressed sequence EH456644 |
| chr1_-_14258409 | 0.39 |
ENSDART00000079359
|
pde5aa
|
phosphodiesterase 5A, cGMP-specific, a |
| chr5_+_37087583 | 0.39 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr5_-_26834511 | 0.39 |
ENSDART00000136713
ENSDART00000192932 ENSDART00000113246 |
si:ch211-102c2.4
|
si:ch211-102c2.4 |
| chr5_-_55623443 | 0.38 |
ENSDART00000005671
ENSDART00000176341 |
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr8_+_11642070 | 0.38 |
ENSDART00000004288
|
ift81
|
intraflagellar transport 81 homolog |
| chr3_+_29942338 | 0.38 |
ENSDART00000158220
|
ifi35
|
interferon-induced protein 35 |
| chr4_-_41209037 | 0.37 |
ENSDART00000151948
|
si:ch211-73m21.1
|
si:ch211-73m21.1 |
| chr13_-_5978433 | 0.37 |
ENSDART00000102555
|
actr2b
|
ARP2 actin related protein 2b homolog |
| chr4_-_64284924 | 0.37 |
ENSDART00000166733
|
CT956002.2
|
|
| chr6_+_40714811 | 0.37 |
ENSDART00000153868
|
ccdc36
|
coiled-coil domain containing 36 |
| chr17_-_681142 | 0.36 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr1_+_22691256 | 0.35 |
ENSDART00000017413
|
zmynd10
|
zinc finger, MYND-type containing 10 |
| chr23_-_45318760 | 0.35 |
ENSDART00000166883
|
ccdc171
|
coiled-coil domain containing 171 |
| chr8_+_8947623 | 0.34 |
ENSDART00000131215
|
slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member 2 |
| chr20_+_42780162 | 0.33 |
ENSDART00000127069
|
pimr213
|
Pim proto-oncogene, serine/threonine kinase, related 213 |
| chr1_-_59407322 | 0.33 |
ENSDART00000161661
|
si:ch211-188p14.5
|
si:ch211-188p14.5 |
| chr10_+_40324395 | 0.33 |
ENSDART00000147205
|
gltpb
|
glycolipid transfer protein b |
| chr19_+_18758014 | 0.32 |
ENSDART00000158935
|
clic1
|
chloride intracellular channel 1 |
| chr24_+_9590188 | 0.32 |
ENSDART00000137092
|
si:dkey-96n2.1
|
si:dkey-96n2.1 |
| chr5_-_23464970 | 0.31 |
ENSDART00000141028
|
si:dkeyp-20g2.1
|
si:dkeyp-20g2.1 |
| chr3_+_46479705 | 0.31 |
ENSDART00000181564
|
tyk2
|
tyrosine kinase 2 |
| chr17_-_19626357 | 0.31 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr8_-_11202378 | 0.31 |
ENSDART00000147817
ENSDART00000174039 |
fam208b
|
family with sequence similarity 208, member B |
| chr21_-_45073764 | 0.31 |
ENSDART00000181390
ENSDART00000063714 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr14_+_6287871 | 0.31 |
ENSDART00000105344
|
tbc1d2
|
TBC1 domain family, member 2 |
| chr3_+_26064091 | 0.31 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr1_-_625875 | 0.31 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr1_-_11104805 | 0.31 |
ENSDART00000147648
|
knl1
|
kinetochore scaffold 1 |
| chr22_+_29067388 | 0.30 |
ENSDART00000133673
|
pimr100
|
Pim proto-oncogene, serine/threonine kinase, related 100 |
| chr1_+_51386649 | 0.29 |
ENSDART00000152289
|
atg4da
|
autophagy related 4D, cysteine peptidase a |
| chr25_-_35664817 | 0.29 |
ENSDART00000148718
|
lrrk2
|
leucine-rich repeat kinase 2 |
| chr3_-_5228137 | 0.29 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr20_+_43602810 | 0.28 |
ENSDART00000142543
|
si:dkey-206f10.1
|
si:dkey-206f10.1 |
| chr23_-_40788933 | 0.27 |
ENSDART00000086613
|
DUPD1 (1 of many)
|
si:dkeyp-27c8.1 |
| chr7_-_24204665 | 0.27 |
ENSDART00000167141
|
gmpr2
|
guanosine monophosphate reductase 2 |
| chr16_+_29514473 | 0.27 |
ENSDART00000034102
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr11_+_13423776 | 0.27 |
ENSDART00000102553
|
homer3b
|
homer scaffolding protein 3b |
| chr2_-_30249746 | 0.27 |
ENSDART00000124153
ENSDART00000142575 |
elocb
|
elongin C paralog b |
| chr3_+_24459709 | 0.27 |
ENSDART00000180976
|
cbx6b
|
chromobox homolog 6b |
| chr11_-_18601955 | 0.26 |
ENSDART00000180565
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr3_-_15080226 | 0.26 |
ENSDART00000109818
ENSDART00000139835 |
nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr17_-_5610514 | 0.26 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr3_+_34234029 | 0.26 |
ENSDART00000044859
|
znf207a
|
zinc finger protein 207, a |
| chr6_-_12459412 | 0.25 |
ENSDART00000090266
ENSDART00000144028 |
gpd2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr5_-_29152457 | 0.24 |
ENSDART00000078469
|
noxa1
|
NADPH oxidase activator 1 |
| chr8_+_50742975 | 0.24 |
ENSDART00000155664
ENSDART00000160612 |
si:ch73-6l19.2
|
si:ch73-6l19.2 |
| chr8_-_41264502 | 0.24 |
ENSDART00000133124
|
rnf10
|
ring finger protein 10 |
| chr24_-_35707552 | 0.23 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr19_-_20403845 | 0.23 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr9_+_2499627 | 0.23 |
ENSDART00000160782
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr3_+_46479913 | 0.22 |
ENSDART00000149755
|
tyk2
|
tyrosine kinase 2 |
| chr6_+_26309968 | 0.22 |
ENSDART00000153805
|
dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr17_+_46739693 | 0.22 |
ENSDART00000097810
|
pimr22
|
Pim proto-oncogene, serine/threonine kinase, related 22 |
| chr4_-_37766174 | 0.22 |
ENSDART00000170066
|
si:dkey-207l24.2
|
si:dkey-207l24.2 |
| chr4_+_2655358 | 0.22 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr23_-_42810664 | 0.22 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr7_-_31759602 | 0.21 |
ENSDART00000113467
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr5_+_40835601 | 0.21 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr15_-_2010926 | 0.21 |
ENSDART00000155688
|
dock10
|
dedicator of cytokinesis 10 |
| chr14_-_26436951 | 0.21 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr12_+_16452912 | 0.21 |
ENSDART00000192467
|
kif20bb
|
kinesin family member 20Bb |
| chr23_+_1181248 | 0.21 |
ENSDART00000170942
|
utrn
|
utrophin |
| chr19_-_19442983 | 0.21 |
ENSDART00000052649
|
sb:cb649
|
sb:cb649 |
| chr3_-_57779801 | 0.21 |
ENSDART00000074285
|
syngr2a
|
synaptogyrin 2a |
| chr9_+_12948511 | 0.21 |
ENSDART00000135797
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr17_-_11418513 | 0.21 |
ENSDART00000064412
ENSDART00000151847 |
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr3_-_34337969 | 0.20 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr1_+_36722122 | 0.20 |
ENSDART00000111566
|
tmem184c
|
transmembrane protein 184C |
| chr1_+_2101541 | 0.20 |
ENSDART00000128187
ENSDART00000167050 ENSDART00000182153 ENSDART00000122626 ENSDART00000164488 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr20_-_22798794 | 0.20 |
ENSDART00000148084
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr4_-_17642168 | 0.20 |
ENSDART00000007030
|
klhl42
|
kelch-like family, member 42 |
| chr18_+_20677090 | 0.20 |
ENSDART00000060243
|
cftr
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr20_-_2090789 | 0.20 |
ENSDART00000113237
|
arhgap18
|
Rho GTPase activating protein 18 |
| chr11_+_34760628 | 0.20 |
ENSDART00000087216
|
si:dkey-202e22.2
|
si:dkey-202e22.2 |
| chr1_+_55293424 | 0.19 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr3_-_5228841 | 0.19 |
ENSDART00000092373
ENSDART00000182438 |
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr6_+_3282809 | 0.19 |
ENSDART00000187444
ENSDART00000187407 ENSDART00000191883 |
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr7_+_22981441 | 0.19 |
ENSDART00000182887
|
ccnb3
|
cyclin B3 |
| chr4_-_17263210 | 0.19 |
ENSDART00000147853
|
lrmp
|
lymphoid-restricted membrane protein |
| chr13_-_44738574 | 0.18 |
ENSDART00000074761
|
zfand3
|
zinc finger, AN1-type domain 3 |
| chr8_+_13694950 | 0.18 |
ENSDART00000132506
|
pimr107
|
Pim proto-oncogene, serine/threonine kinase, related 107 |
| chr8_+_35964482 | 0.18 |
ENSDART00000129357
ENSDART00000154953 |
glt1d1
|
glycosyltransferase 1 domain containing 1 |
| chr15_-_1534232 | 0.18 |
ENSDART00000056763
ENSDART00000133943 |
ift80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr1_+_10318089 | 0.18 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr3_+_57997980 | 0.18 |
ENSDART00000168477
ENSDART00000193840 |
pycr1a
|
pyrroline-5-carboxylate reductase 1a |
| chr22_+_29009541 | 0.18 |
ENSDART00000169449
|
pimr97
|
Pim proto-oncogene, serine/threonine kinase, related 97 |
| chr11_+_45343245 | 0.18 |
ENSDART00000160904
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr2_+_27651984 | 0.18 |
ENSDART00000126762
|
tmem68
|
transmembrane protein 68 |
| chr8_-_13315304 | 0.18 |
ENSDART00000142596
|
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr9_-_49860756 | 0.18 |
ENSDART00000044270
|
ttc21b
|
tetratricopeptide repeat domain 21B |
| chr7_-_31759394 | 0.17 |
ENSDART00000193040
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr12_+_34763795 | 0.17 |
ENSDART00000153097
|
slc38a10
|
solute carrier family 38, member 10 |
| chr19_+_25476228 | 0.17 |
ENSDART00000112808
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr21_+_25198637 | 0.17 |
ENSDART00000164972
|
si:dkey-183i3.6
|
si:dkey-183i3.6 |
| chr1_+_55294009 | 0.17 |
ENSDART00000128979
|
zgc:172106
|
zgc:172106 |
| chr8_+_15239549 | 0.17 |
ENSDART00000132216
|
paox1
|
polyamine oxidase (exo-N4-amino) 1 |
| chr5_-_39171302 | 0.17 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr4_-_17353100 | 0.16 |
ENSDART00000134467
ENSDART00000189019 |
parpbp
|
PARP1 binding protein |
| chr13_+_34689663 | 0.16 |
ENSDART00000133661
|
tasp1
|
taspase, threonine aspartase, 1 |
| chr8_+_53423408 | 0.16 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr13_+_27316934 | 0.16 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr7_+_31051603 | 0.16 |
ENSDART00000108721
|
tjp1a
|
tight junction protein 1a |
| chr15_-_8856391 | 0.16 |
ENSDART00000008273
|
rab4b
|
RAB4B, member RAS oncogene family |
| chr25_-_32311048 | 0.16 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr1_-_47071979 | 0.15 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr11_-_141592 | 0.15 |
ENSDART00000092787
|
cdk4
|
cyclin-dependent kinase 4 |
| chr12_+_33395748 | 0.15 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr4_+_14727212 | 0.15 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr6_-_442163 | 0.15 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr2_+_27652386 | 0.15 |
ENSDART00000188261
|
tmem68
|
transmembrane protein 68 |
| chr1_-_24349759 | 0.15 |
ENSDART00000142740
ENSDART00000177989 |
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr2_+_17524278 | 0.15 |
ENSDART00000165633
|
pimr196
|
Pim proto-oncogene, serine/threonine kinase, related 196 |
| chr7_+_19374683 | 0.15 |
ENSDART00000162700
|
snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr15_+_21882419 | 0.14 |
ENSDART00000157216
|
si:dkey-103g5.4
|
si:dkey-103g5.4 |
| chr5_-_31875645 | 0.14 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr12_-_7824291 | 0.14 |
ENSDART00000148673
ENSDART00000149453 |
ank3b
|
ankyrin 3b |
| chr6_-_8489810 | 0.14 |
ENSDART00000124643
|
rasal3
|
RAS protein activator like 3 |
| chr5_+_65946222 | 0.14 |
ENSDART00000190969
ENSDART00000161578 |
mymk
|
myomaker, myoblast fusion factor |
| chr23_+_42304602 | 0.14 |
ENSDART00000166004
|
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr15_+_9327252 | 0.14 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr22_-_30973791 | 0.14 |
ENSDART00000104728
|
ssuh2.2
|
ssu-2 homolog, tandem duplicate 2 |
| chr4_-_9533345 | 0.14 |
ENSDART00000128501
|
si:ch73-139j3.4
|
si:ch73-139j3.4 |
| chr7_+_22981909 | 0.13 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr11_-_29816429 | 0.13 |
ENSDART00000069132
|
cybb
|
cytochrome b-245, beta polypeptide (chronic granulomatous disease) |
| chr3_-_9458445 | 0.13 |
ENSDART00000192712
|
FO904885.1
|
|
| chr3_+_24612191 | 0.13 |
ENSDART00000190998
|
zgc:171506
|
zgc:171506 |
| chr22_-_3595439 | 0.13 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr7_-_6351021 | 0.13 |
ENSDART00000159542
|
zgc:112234
|
zgc:112234 |
| chr20_+_32478151 | 0.13 |
ENSDART00000145269
|
ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr1_+_12767318 | 0.13 |
ENSDART00000162652
|
pcdh10a
|
protocadherin 10a |
| chr20_-_33556983 | 0.13 |
ENSDART00000168798
|
rock2bl
|
rho-associated, coiled-coil containing protein kinase 2b, like |
| chr16_-_12723738 | 0.13 |
ENSDART00000080414
|
sbk3
|
SH3 domain binding kinase family, member 3 |
| chr4_-_12723585 | 0.13 |
ENSDART00000185639
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr24_+_37449893 | 0.13 |
ENSDART00000164405
|
nlrc3
|
NLR family, CARD domain containing 3 |
| chr10_-_26766780 | 0.13 |
ENSDART00000146666
|
mcf2b
|
MCF.2 cell line derived transforming sequence b |
| chr22_+_1170294 | 0.13 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr12_-_14551077 | 0.13 |
ENSDART00000188717
|
FO704673.1
|
|
| chr8_+_22438398 | 0.12 |
ENSDART00000135145
|
zgc:153759
|
zgc:153759 |
| chr13_+_28705143 | 0.12 |
ENSDART00000183338
|
ldb1a
|
LIM domain binding 1a |
| chr21_+_21244046 | 0.12 |
ENSDART00000058306
ENSDART00000189665 ENSDART00000131890 |
lifrb
|
leukemia inhibitory factor receptor alpha b |
| chr1_-_35924495 | 0.12 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr8_-_46321889 | 0.12 |
ENSDART00000075189
ENSDART00000122801 |
mtor
|
mechanistic target of rapamycin kinase |
| chr8_+_48801402 | 0.12 |
ENSDART00000098363
|
tprg1l
|
tumor protein p63 regulated 1-like |
| chr15_+_15856178 | 0.12 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr21_-_2217685 | 0.12 |
ENSDART00000159315
|
si:dkey-50i6.5
|
si:dkey-50i6.5 |
| chr6_+_18298444 | 0.12 |
ENSDART00000166018
|
card14
|
caspase recruitment domain family, member 14 |
| chr3_-_50277959 | 0.12 |
ENSDART00000082773
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr24_-_11069237 | 0.12 |
ENSDART00000127398
|
CR753886.1
|
|
| chr11_-_38914265 | 0.12 |
ENSDART00000141229
|
si:ch211-122l14.4
|
si:ch211-122l14.4 |
| chr15_+_15415623 | 0.11 |
ENSDART00000127436
|
zgc:92630
|
zgc:92630 |
| chr3_-_21061931 | 0.11 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr1_+_157793 | 0.11 |
ENSDART00000152205
|
cul4a
|
cullin 4A |
| chr19_+_48373140 | 0.11 |
ENSDART00000160896
|
abhd5b
|
abhydrolase domain containing 5b |
| chr17_+_24222190 | 0.11 |
ENSDART00000181698
ENSDART00000189411 |
ehbp1
|
EH domain binding protein 1 |
| chr15_+_1534644 | 0.11 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr15_+_3808996 | 0.11 |
ENSDART00000110227
|
RNF14 (1 of many)
|
ring finger protein 14 |
| chr4_+_9911534 | 0.11 |
ENSDART00000109452
|
sbf1
|
SET binding factor 1 |
| chr16_+_13855039 | 0.11 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr7_+_35075847 | 0.11 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr7_-_56766973 | 0.11 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr25_-_34979548 | 0.10 |
ENSDART00000153904
|
si:dkey-108k21.7
|
si:dkey-108k21.7 |
| chr4_-_12978925 | 0.10 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr4_-_22310956 | 0.10 |
ENSDART00000162585
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr23_-_9965148 | 0.10 |
ENSDART00000137308
|
plxnb1a
|
plexin b1a |
| chr3_-_11972516 | 0.10 |
ENSDART00000140123
|
hmox2b
|
heme oxygenase 2b |
| chr22_-_16377960 | 0.10 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr24_-_32025637 | 0.10 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr13_+_30696286 | 0.10 |
ENSDART00000192411
|
cxcl18a.1
|
chemokine (C-X-C motif) ligand 18a, duplicate 1 |
| chr15_-_8856785 | 0.10 |
ENSDART00000192816
|
rab4b
|
RAB4B, member RAS oncogene family |
| chr20_-_33705044 | 0.10 |
ENSDART00000166573
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of elf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.2 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.1 | 0.4 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.4 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.4 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.2 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.6 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.3 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.1 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.0 | 0.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.4 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.6 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.3 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0002902 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 | 0.3 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.0 | GO:0045191 | regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.3 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 5.8 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.0 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.1 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.5 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.1 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.0 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.0 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.3 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |