Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
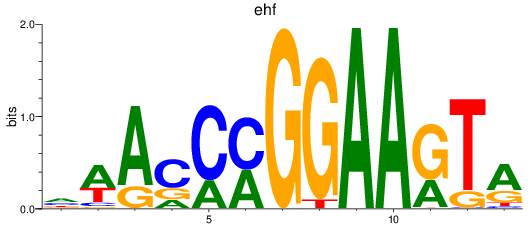
Results for ehf
Z-value: 0.58
Transcription factors associated with ehf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ehf
|
ENSDARG00000052115 | ets homologous factor |
|
ehf
|
ENSDARG00000112589 | ets homologous factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ehf | dr11_v1_chr25_+_10416583_10416583 | -0.54 | 2.0e-02 | Click! |
Activity profile of ehf motif
Sorted Z-values of ehf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_40237276 | 1.88 |
ENSDART00000037553
|
faah2a
|
fatty acid amide hydrolase 2a |
| chr5_+_9134187 | 1.79 |
ENSDART00000132240
|
pimr123
|
Pim proto-oncogene, serine/threonine kinase, related 123 |
| chr9_-_48397702 | 1.79 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr16_-_33930759 | 1.77 |
ENSDART00000177453
|
dnali1
|
dynein, axonemal, light intermediate chain 1 |
| chr5_-_57920600 | 1.74 |
ENSDART00000132376
|
si:dkey-27p23.3
|
si:dkey-27p23.3 |
| chr24_+_17269849 | 1.70 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr1_+_55239160 | 1.68 |
ENSDART00000152318
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr10_+_40324395 | 1.64 |
ENSDART00000147205
|
gltpb
|
glycolipid transfer protein b |
| chr14_+_21007957 | 1.64 |
ENSDART00000162316
|
pimr194
|
Pim proto-oncogene, serine/threonine kinase, related 194 |
| chr11_-_12512122 | 1.59 |
ENSDART00000145338
|
si:dkey-27d5.3
|
si:dkey-27d5.3 |
| chr8_+_17184602 | 1.57 |
ENSDART00000050228
ENSDART00000140531 |
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr10_-_5854862 | 1.56 |
ENSDART00000162813
|
si:ch211-281k19.2
|
si:ch211-281k19.2 |
| chr5_+_32260502 | 1.41 |
ENSDART00000149020
|
si:ch211-158m24.12
|
si:ch211-158m24.12 |
| chr8_+_17987778 | 1.40 |
ENSDART00000133429
|
lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr4_-_5826320 | 1.40 |
ENSDART00000165354
|
foxm1
|
forkhead box M1 |
| chr20_-_43572763 | 1.39 |
ENSDART00000153251
|
pimr125
|
Pim proto-oncogene, serine/threonine kinase, related 125 |
| chr12_-_2993095 | 1.36 |
ENSDART00000152316
|
si:dkey-202c14.3
|
si:dkey-202c14.3 |
| chr6_-_40352215 | 1.36 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr17_-_10309766 | 1.35 |
ENSDART00000160994
|
ttc6
|
tetratricopeptide repeat domain 6 |
| chr24_+_17270129 | 1.31 |
ENSDART00000186729
|
spag6
|
sperm associated antigen 6 |
| chr2_+_9061885 | 1.30 |
ENSDART00000028906
|
pigk
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr8_+_17987215 | 1.28 |
ENSDART00000113605
|
lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr2_-_11662851 | 1.27 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr8_+_15239549 | 1.26 |
ENSDART00000132216
|
paox1
|
polyamine oxidase (exo-N4-amino) 1 |
| chr5_+_9112291 | 1.25 |
ENSDART00000135177
|
AL845369.1
|
|
| chr4_-_13567387 | 1.25 |
ENSDART00000132971
ENSDART00000102010 |
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr7_+_19374683 | 1.24 |
ENSDART00000162700
|
snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr11_-_38899816 | 1.23 |
ENSDART00000141515
|
pimr127
|
Pim proto-oncogene, serine/threonine kinase, related 127 |
| chr14_+_21042178 | 1.21 |
ENSDART00000168551
|
pimr192
|
Pim proto-oncogene, serine/threonine kinase, related 192 |
| chr6_+_18251140 | 1.21 |
ENSDART00000169752
|
ccdc40
|
coiled-coil domain containing 40 |
| chr24_+_9590188 | 1.17 |
ENSDART00000137092
|
si:dkey-96n2.1
|
si:dkey-96n2.1 |
| chr8_-_6918721 | 1.17 |
ENSDART00000014915
|
asb6
|
ankyrin repeat and SOCS box containing 6 |
| chr20_+_39457598 | 1.13 |
ENSDART00000140931
ENSDART00000156176 |
pimr128
|
Pim proto-oncogene, serine/threonine kinase, related 128 |
| chr1_+_55239710 | 1.12 |
ENSDART00000174846
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr8_-_6918290 | 1.12 |
ENSDART00000138259
ENSDART00000142496 |
asb6
|
ankyrin repeat and SOCS box containing 6 |
| chr24_-_24959607 | 1.11 |
ENSDART00000146930
ENSDART00000184482 |
pdk3a
|
pyruvate dehydrogenase kinase, isozyme 3a |
| chr4_+_842010 | 1.10 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr11_-_12219829 | 1.09 |
ENSDART00000168025
ENSDART00000112677 ENSDART00000108681 |
si:ch211-156l18.6
|
si:ch211-156l18.6 |
| chr21_+_28502340 | 1.07 |
ENSDART00000077897
ENSDART00000140229 |
otub1a
|
OTU deubiquitinase, ubiquitin aldehyde binding 1a |
| chr3_-_18030938 | 1.07 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr10_+_6613279 | 1.06 |
ENSDART00000149643
|
si:ch211-57m13.1
|
si:ch211-57m13.1 |
| chr20_+_42780162 | 1.05 |
ENSDART00000127069
|
pimr213
|
Pim proto-oncogene, serine/threonine kinase, related 213 |
| chr19_+_31873308 | 1.05 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr20_-_43495506 | 1.03 |
ENSDART00000170679
|
pimr132
|
Pim proto-oncogene, serine/threonine kinase, related 132 |
| chr20_+_42761881 | 1.00 |
ENSDART00000113625
|
pimr113
|
Pim proto-oncogene, serine/threonine kinase, related 113 |
| chr10_+_26667475 | 0.98 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr14_+_81919 | 0.97 |
ENSDART00000171444
|
stag3
|
stromal antigen 3 |
| chr2_-_14272083 | 0.95 |
ENSDART00000169986
|
tctex1d1
|
Tctex1 domain containing 1 |
| chr20_-_43462190 | 0.95 |
ENSDART00000100697
|
pimr133
|
Pim proto-oncogene, serine/threonine kinase, related 133 |
| chr7_-_20103384 | 0.93 |
ENSDART00000052902
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr22_+_2937485 | 0.91 |
ENSDART00000082222
ENSDART00000135507 ENSDART00000143258 |
cep19
|
centrosomal protein 19 |
| chr23_+_20687340 | 0.91 |
ENSDART00000143503
|
usp21
|
ubiquitin specific peptidase 21 |
| chr6_-_12459412 | 0.91 |
ENSDART00000090266
ENSDART00000144028 |
gpd2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr6_+_49095646 | 0.90 |
ENSDART00000103385
|
slc25a55a
|
solute carrier family 25, member 55a |
| chr6_-_21988375 | 0.90 |
ENSDART00000161257
|
plxnb1b
|
plexin b1b |
| chr13_-_36703164 | 0.89 |
ENSDART00000044357
|
cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr25_-_16969922 | 0.87 |
ENSDART00000111158
|
tigara
|
tp53-induced glycolysis and apoptosis regulator a |
| chr1_+_22691256 | 0.87 |
ENSDART00000017413
|
zmynd10
|
zinc finger, MYND-type containing 10 |
| chr20_-_43602650 | 0.86 |
ENSDART00000152939
|
pimr124
|
Pim proto-oncogene, serine/threonine kinase, related 124 |
| chr9_+_38712474 | 0.86 |
ENSDART00000182874
|
kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr19_+_17385561 | 0.85 |
ENSDART00000141397
ENSDART00000143913 ENSDART00000133626 |
rpl15
|
ribosomal protein L15 |
| chr6_+_11990733 | 0.85 |
ENSDART00000151075
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr24_+_40473032 | 0.84 |
ENSDART00000084238
ENSDART00000178508 |
CABZ01076968.1
|
|
| chr11_-_12394682 | 0.84 |
ENSDART00000172447
|
si:dkey-27d5.10
|
si:dkey-27d5.10 |
| chr3_+_36424055 | 0.84 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr23_-_19140781 | 0.84 |
ENSDART00000143580
|
si:ch73-381f5.2
|
si:ch73-381f5.2 |
| chr4_-_41209037 | 0.83 |
ENSDART00000151948
|
si:ch211-73m21.1
|
si:ch211-73m21.1 |
| chr21_-_37733571 | 0.83 |
ENSDART00000176214
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr8_+_11642070 | 0.83 |
ENSDART00000004288
|
ift81
|
intraflagellar transport 81 homolog |
| chr1_+_49510209 | 0.82 |
ENSDART00000052998
|
si:dkeyp-80c12.4
|
si:dkeyp-80c12.4 |
| chr11_+_2642013 | 0.81 |
ENSDART00000111324
|
zgc:193807
|
zgc:193807 |
| chr14_-_30724165 | 0.81 |
ENSDART00000020936
|
fibpa
|
fibroblast growth factor (acidic) intracellular binding protein a |
| chr16_-_41059329 | 0.81 |
ENSDART00000145556
|
si:dkey-201i6.8
|
si:dkey-201i6.8 |
| chr18_+_2168695 | 0.80 |
ENSDART00000165855
|
dnaaf4
|
dynein axonemal assembly factor 4 |
| chr7_+_22823889 | 0.80 |
ENSDART00000127467
ENSDART00000148576 ENSDART00000149993 |
pygmb
|
phosphorylase, glycogen, muscle b |
| chr20_+_39416729 | 0.79 |
ENSDART00000099993
ENSDART00000144133 |
pimr128
|
Pim proto-oncogene, serine/threonine kinase, related 128 |
| chr5_-_34616599 | 0.79 |
ENSDART00000050271
ENSDART00000097975 |
hexb
|
hexosaminidase B (beta polypeptide) |
| chr2_-_14271874 | 0.77 |
ENSDART00000189390
|
tctex1d1
|
Tctex1 domain containing 1 |
| chr17_-_13026634 | 0.77 |
ENSDART00000113713
|
fam177a1
|
family with sequence similarity 177, member A1 |
| chr11_+_8129536 | 0.77 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr5_+_4366431 | 0.76 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr17_-_24684687 | 0.76 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr2_+_31597841 | 0.75 |
ENSDART00000162181
|
si:ch211-106h4.4
|
si:ch211-106h4.4 |
| chr11_-_12499116 | 0.75 |
ENSDART00000134836
|
si:dkey-27d5.4
|
si:dkey-27d5.4 |
| chr1_-_17711636 | 0.75 |
ENSDART00000148322
ENSDART00000122670 |
ufsp2
|
ufm1-specific peptidase 2 |
| chr12_-_20795867 | 0.74 |
ENSDART00000152835
ENSDART00000153424 |
nme2a
|
NME/NM23 nucleoside diphosphate kinase 2a |
| chr23_+_19182819 | 0.74 |
ENSDART00000131804
|
si:dkey-93l1.4
|
si:dkey-93l1.4 |
| chr21_-_8153165 | 0.74 |
ENSDART00000182580
|
CABZ01074363.1
|
|
| chr23_+_19691146 | 0.73 |
ENSDART00000143001
|
si:dkey-93l1.6
|
si:dkey-93l1.6 |
| chr2_-_30693742 | 0.73 |
ENSDART00000090292
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr24_-_982443 | 0.73 |
ENSDART00000063151
|
napga
|
N-ethylmaleimide-sensitive factor attachment protein, gamma a |
| chr8_+_39760258 | 0.73 |
ENSDART00000037914
|
cox6a1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr11_-_12350419 | 0.72 |
ENSDART00000166732
|
si:dkey-27d5.14
|
si:dkey-27d5.14 |
| chr16_+_8668519 | 0.72 |
ENSDART00000172893
|
ccr2
|
chemokine (C-C motif) receptor 2 |
| chr20_-_26551210 | 0.71 |
ENSDART00000077715
|
si:dkey-25e12.3
|
si:dkey-25e12.3 |
| chr20_+_7584211 | 0.70 |
ENSDART00000132481
ENSDART00000127975 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr20_+_39394308 | 0.69 |
ENSDART00000142554
|
pimr136
|
Pim proto-oncogene, serine/threonine kinase, related 136 |
| chr13_-_5978433 | 0.68 |
ENSDART00000102555
|
actr2b
|
ARP2 actin related protein 2b homolog |
| chr1_-_47161996 | 0.68 |
ENSDART00000053153
|
mhc1zba
|
major histocompatibility complex class I ZBA |
| chr22_-_16042243 | 0.68 |
ENSDART00000062633
|
s1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr13_-_24311628 | 0.68 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr8_-_52715911 | 0.67 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr16_-_41867833 | 0.67 |
ENSDART00000133066
|
zgc:194418
|
zgc:194418 |
| chr1_+_2101541 | 0.67 |
ENSDART00000128187
ENSDART00000167050 ENSDART00000182153 ENSDART00000122626 ENSDART00000164488 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr20_-_16223245 | 0.67 |
ENSDART00000014721
|
echdc2
|
enoyl CoA hydratase domain containing 2 |
| chr1_-_14258409 | 0.67 |
ENSDART00000079359
|
pde5aa
|
phosphodiesterase 5A, cGMP-specific, a |
| chr1_+_9199031 | 0.66 |
ENSDART00000092058
ENSDART00000182771 |
chtf18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
| chr22_+_16320076 | 0.66 |
ENSDART00000164161
|
osbpl1a
|
oxysterol binding protein-like 1A |
| chr12_-_47793857 | 0.66 |
ENSDART00000161294
|
dydc2
|
DPY30 domain containing 2 |
| chr16_+_25316973 | 0.66 |
ENSDART00000086409
|
dync1i1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr19_-_42424599 | 0.65 |
ENSDART00000077042
|
zgc:153441
|
zgc:153441 |
| chr6_+_6797520 | 0.65 |
ENSDART00000150967
|
si:ch211-85n16.3
|
si:ch211-85n16.3 |
| chr10_-_36726188 | 0.65 |
ENSDART00000157262
|
pimr137
|
Pim proto-oncogene, serine/threonine kinase, related 137 |
| chr7_-_72352043 | 0.65 |
ENSDART00000163536
|
muc5f
|
mucin 5f |
| chr6_-_41085443 | 0.65 |
ENSDART00000143944
ENSDART00000028217 |
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr5_+_37087583 | 0.64 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr14_+_28438947 | 0.63 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long chain family member 4a |
| chr25_-_37435060 | 0.63 |
ENSDART00000102855
ENSDART00000148566 |
pdhx
|
pyruvate dehydrogenase complex, component X |
| chr3_+_40809011 | 0.62 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr11_+_8660158 | 0.62 |
ENSDART00000169141
|
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr25_+_36045072 | 0.61 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr23_-_40788933 | 0.61 |
ENSDART00000086613
|
DUPD1 (1 of many)
|
si:dkeyp-27c8.1 |
| chr15_-_23793641 | 0.60 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr6_+_10871659 | 0.60 |
ENSDART00000041858
|
mtx2
|
metaxin 2 |
| chr17_+_24613255 | 0.60 |
ENSDART00000064738
|
atp5if1b
|
ATP synthase inhibitory factor subunit 1b |
| chr20_+_43602810 | 0.59 |
ENSDART00000142543
|
si:dkey-206f10.1
|
si:dkey-206f10.1 |
| chr11_-_38928760 | 0.59 |
ENSDART00000146277
|
si:ch211-122l14.6
|
si:ch211-122l14.6 |
| chr25_-_37489917 | 0.59 |
ENSDART00000160688
|
psma1
|
proteasome subunit alpha 1 |
| chr19_+_11978209 | 0.59 |
ENSDART00000111568
|
polr2k
|
polymerase (RNA) II (DNA directed) polypeptide K |
| chr7_+_65673885 | 0.59 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr18_+_22994113 | 0.58 |
ENSDART00000171871
|
cbfb
|
core-binding factor, beta subunit |
| chr17_+_31611854 | 0.58 |
ENSDART00000011706
|
adprhl2
|
ADP-ribosylhydrolase like 2 |
| chr17_-_20218357 | 0.58 |
ENSDART00000155990
ENSDART00000155632 ENSDART00000156540 ENSDART00000063523 |
mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr5_+_19867794 | 0.57 |
ENSDART00000051614
|
tchp
|
trichoplein, keratin filament binding |
| chr2_-_42415902 | 0.57 |
ENSDART00000142489
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr20_+_39373480 | 0.56 |
ENSDART00000136714
|
pimr131
|
Pim proto-oncogene, serine/threonine kinase, related 131 |
| chr12_-_14922955 | 0.56 |
ENSDART00000002078
|
neurod2
|
neurogenic differentiation 2 |
| chr14_-_24391424 | 0.56 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr3_+_26064091 | 0.55 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr8_+_54284961 | 0.55 |
ENSDART00000122692
|
plxnd1
|
plexin D1 |
| chr23_-_45022681 | 0.54 |
ENSDART00000102640
|
ctc1
|
CTS telomere maintenance complex component 1 |
| chr22_-_26865361 | 0.53 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr17_+_24809221 | 0.52 |
ENSDART00000082251
ENSDART00000147871 ENSDART00000130871 |
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr19_-_48391415 | 0.52 |
ENSDART00000170726
ENSDART00000169577 |
si:ch73-359m17.2
|
si:ch73-359m17.2 |
| chr7_-_59054322 | 0.52 |
ENSDART00000165390
|
chmp5b
|
charged multivesicular body protein 5b |
| chr12_+_16452575 | 0.52 |
ENSDART00000058665
|
kif20bb
|
kinesin family member 20Bb |
| chr3_-_32902138 | 0.52 |
ENSDART00000144026
ENSDART00000083874 ENSDART00000145443 ENSDART00000148239 ENSDART00000134917 |
kat7a
|
K(lysine) acetyltransferase 7a |
| chr13_-_32626247 | 0.52 |
ENSDART00000100663
|
wdr35
|
WD repeat domain 35 |
| chr20_+_54349333 | 0.52 |
ENSDART00000132168
ENSDART00000147542 |
znf410
|
zinc finger protein 410 |
| chr16_-_41439659 | 0.52 |
ENSDART00000191624
|
cpne4a
|
copine IVa |
| chr6_-_29288155 | 0.51 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr9_-_49860756 | 0.51 |
ENSDART00000044270
|
ttc21b
|
tetratricopeptide repeat domain 21B |
| chr14_-_28567845 | 0.51 |
ENSDART00000126095
|
insb
|
preproinsulin b |
| chr16_+_16968682 | 0.51 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr13_+_27316934 | 0.51 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr20_+_22666548 | 0.50 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr17_-_49995366 | 0.50 |
ENSDART00000075223
|
cox7a2a
|
cytochrome c oxidase subunit VIIa polypeptide 2a (liver) |
| chr1_+_55294009 | 0.50 |
ENSDART00000128979
|
zgc:172106
|
zgc:172106 |
| chr9_-_27857265 | 0.50 |
ENSDART00000177513
ENSDART00000143166 ENSDART00000115313 |
si:rp71-45g20.4
|
si:rp71-45g20.4 |
| chr14_-_28568107 | 0.49 |
ENSDART00000042850
ENSDART00000145502 |
insb
|
preproinsulin b |
| chr9_-_31747106 | 0.49 |
ENSDART00000048469
ENSDART00000145204 ENSDART00000186889 |
nalcn
|
sodium leak channel, non-selective |
| chr22_-_26865181 | 0.49 |
ENSDART00000138311
|
hmox2a
|
heme oxygenase 2a |
| chr16_-_45001842 | 0.49 |
ENSDART00000037797
|
sult2st3
|
sulfotransferase family 2, cytosolic sulfotransferase 3 |
| chr17_-_20218167 | 0.49 |
ENSDART00000154479
|
mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr16_-_52821023 | 0.48 |
ENSDART00000074718
|
spire1b
|
spire-type actin nucleation factor 1b |
| chr10_+_26571174 | 0.48 |
ENSDART00000148617
ENSDART00000112956 |
slc9a6b
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b |
| chr12_+_34763795 | 0.47 |
ENSDART00000153097
|
slc38a10
|
solute carrier family 38, member 10 |
| chr4_-_149334 | 0.47 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr12_+_25097754 | 0.47 |
ENSDART00000050070
|
pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr21_+_723998 | 0.47 |
ENSDART00000160956
|
oaz1b
|
ornithine decarboxylase antizyme 1b |
| chr8_-_17167819 | 0.47 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr16_-_48673938 | 0.46 |
ENSDART00000156969
|
notchl
|
notch homolog, like |
| chr10_-_5016997 | 0.46 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr9_-_22821901 | 0.46 |
ENSDART00000101711
|
neb
|
nebulin |
| chr3_-_40945710 | 0.46 |
ENSDART00000138719
ENSDART00000102416 |
cyp3c4
|
cytochrome P450, family 3, subfamily C, polypeptide 4 |
| chr8_+_2757821 | 0.46 |
ENSDART00000051403
ENSDART00000160551 |
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr4_-_22311610 | 0.46 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr6_-_42049350 | 0.46 |
ENSDART00000022949
|
parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr13_+_28819768 | 0.46 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr23_+_27675581 | 0.45 |
ENSDART00000127198
|
rps26
|
ribosomal protein S26 |
| chr7_+_48805534 | 0.45 |
ENSDART00000145375
ENSDART00000148744 |
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr1_+_55293424 | 0.45 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr4_+_52347226 | 0.45 |
ENSDART00000171626
|
si:dkeyp-107f9.2
|
si:dkeyp-107f9.2 |
| chr5_-_57480660 | 0.44 |
ENSDART00000147875
ENSDART00000142776 |
si:ch211-202f5.2
|
si:ch211-202f5.2 |
| chr20_-_32188897 | 0.44 |
ENSDART00000133887
|
si:ch211-51a19.5
|
si:ch211-51a19.5 |
| chr4_+_52162176 | 0.44 |
ENSDART00000167023
|
si:dkeyp-107f9.1
|
si:dkeyp-107f9.1 |
| chr24_-_6078222 | 0.44 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr16_+_53489676 | 0.44 |
ENSDART00000074653
|
grinab
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1b (glutamate binding) |
| chr18_+_28988373 | 0.44 |
ENSDART00000018685
|
syt9a
|
synaptotagmin IXa |
| chr2_-_9818640 | 0.44 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr25_+_5068442 | 0.44 |
ENSDART00000097522
|
parvg
|
parvin, gamma |
| chr19_-_11977783 | 0.44 |
ENSDART00000183337
|
si:ch1073-296d18.1
|
si:ch1073-296d18.1 |
| chr7_-_19923249 | 0.43 |
ENSDART00000078694
|
zgc:110591
|
zgc:110591 |
| chr9_-_42484444 | 0.43 |
ENSDART00000048320
ENSDART00000047653 |
tfpia
|
tissue factor pathway inhibitor a |
| chr5_+_54400971 | 0.43 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr12_-_46985 | 0.43 |
ENSDART00000152327
|
wdr45b
|
WD repeat domain 45B |
| chr17_-_38778826 | 0.43 |
ENSDART00000168182
ENSDART00000124041 ENSDART00000136921 |
dglucy
|
D-glutamate cyclase |
| chr12_+_16452912 | 0.43 |
ENSDART00000192467
|
kif20bb
|
kinesin family member 20Bb |
| chr13_+_28705143 | 0.43 |
ENSDART00000183338
|
ldb1a
|
LIM domain binding 1a |
| chr7_+_26172396 | 0.43 |
ENSDART00000173975
|
si:ch211-196f2.7
|
si:ch211-196f2.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ehf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.3 | 0.9 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.3 | 0.8 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.2 | 1.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 0.7 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 0.8 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.2 | 0.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 0.7 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.2 | 1.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 1.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 0.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 1.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 1.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.6 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.4 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) regulation of apoptotic cell clearance(GO:2000425) |
| 0.1 | 0.8 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.4 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.5 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 0.3 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.1 | 0.6 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.1 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 1.4 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.1 | 0.5 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 1.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 1.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.4 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.9 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.6 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.7 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.8 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.7 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.4 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.6 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 1.4 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.2 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 1.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.6 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.5 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.4 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.6 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.5 | GO:0009118 | regulation of oxidative phosphorylation(GO:0002082) regulation of nucleoside metabolic process(GO:0009118) regulation of ATP metabolic process(GO:1903578) |
| 0.1 | 0.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 20.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 1.1 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.3 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.3 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.3 | GO:0097638 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.5 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.9 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 1.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.4 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.4 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0048913 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.5 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.0 | 0.8 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.1 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.0 | 0.4 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.5 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.4 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.6 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.7 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:1903059 | regulation of lipoprotein metabolic process(GO:0050746) regulation of protein lipidation(GO:1903059) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.5 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.2 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.5 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 1.2 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:1905208 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 0.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.2 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.2 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.0 | 0.1 | GO:0036260 | 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 | 0.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.1 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.5 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.7 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.3 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.7 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 1.0 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.4 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0009154 | purine ribonucleotide catabolic process(GO:0009154) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:1902623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 0.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.7 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.8 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.6 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.6 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.9 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 1.2 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.8 | GO:0030990 | intraciliary transport particle(GO:0030990) intraciliary transport particle B(GO:0030992) |
| 0.0 | 3.1 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.0 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.4 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 2.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 2.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.0 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 1.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.3 | 1.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 1.6 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.3 | 1.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.2 | 1.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 0.7 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.2 | 1.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 0.5 | GO:0009013 | succinate-semialdehyde dehydrogenase (NAD+) activity(GO:0004777) succinate-semialdehyde dehydrogenase [NAD(P)+] activity(GO:0009013) |
| 0.2 | 0.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.7 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 1.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 1.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.4 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 2.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.4 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 0.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.9 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.8 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.1 | 0.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 0.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.6 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.1 | 0.4 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.6 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.3 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.9 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.9 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.8 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 1.2 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 1.1 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.0 | 0.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 16.1 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 3.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.8 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 1.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 2.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |