Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
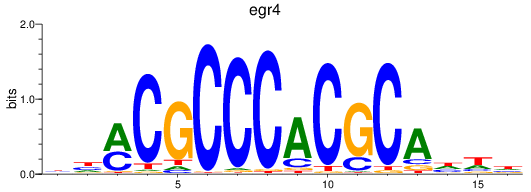
Results for egr4
Z-value: 0.29
Transcription factors associated with egr4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr4
|
ENSDARG00000077799 | early growth response 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr4 | dr11_v1_chr23_+_45579497_45579497 | 0.62 | 5.6e-03 | Click! |
Activity profile of egr4 motif
Sorted Z-values of egr4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_30298377 | 0.53 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr3_-_32180796 | 0.51 |
ENSDART00000133191
ENSDART00000055308 |
pih1d1
|
PIH1 domain containing 1 |
| chr24_+_24831727 | 0.45 |
ENSDART00000080969
|
trim55b
|
tripartite motif containing 55b |
| chr16_-_12784373 | 0.43 |
ENSDART00000080396
|
foxj2
|
forkhead box J2 |
| chr13_-_44834895 | 0.41 |
ENSDART00000135573
|
si:dkeyp-2e4.5
|
si:dkeyp-2e4.5 |
| chr2_+_16710889 | 0.39 |
ENSDART00000017852
|
ubxn7
|
UBX domain protein 7 |
| chr16_-_54405976 | 0.38 |
ENSDART00000055395
|
osr2
|
odd-skipped related transciption factor 2 |
| chr16_-_40508624 | 0.37 |
ENSDART00000075718
|
ndufaf6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr23_+_39606108 | 0.37 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr4_+_842010 | 0.36 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr13_-_36418921 | 0.35 |
ENSDART00000135804
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr12_-_23129453 | 0.32 |
ENSDART00000077453
|
armc4
|
armadillo repeat containing 4 |
| chr2_-_48753873 | 0.28 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr18_+_20481982 | 0.28 |
ENSDART00000128139
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr11_-_165288 | 0.28 |
ENSDART00000108703
ENSDART00000173151 |
tegt
|
testis enhanced gene transcript (BAX inhibitor 1) |
| chr18_+_20482369 | 0.28 |
ENSDART00000100668
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr3_+_31897029 | 0.27 |
ENSDART00000146275
|
strada
|
ste20-related kinase adaptor alpha |
| chr21_-_31238244 | 0.26 |
ENSDART00000159678
|
tpst1l
|
tyrosylprotein sulfotransferase 1, like |
| chr17_-_10025234 | 0.26 |
ENSDART00000008355
|
cfl2
|
cofilin 2 (muscle) |
| chr17_-_2584423 | 0.25 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr21_-_41838284 | 0.23 |
ENSDART00000141067
|
cct6a
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr11_-_236984 | 0.23 |
ENSDART00000170778
|
dusp7
|
dual specificity phosphatase 7 |
| chr2_+_105748 | 0.22 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr18_-_45617146 | 0.21 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr11_-_236766 | 0.21 |
ENSDART00000163978
|
dusp7
|
dual specificity phosphatase 7 |
| chr7_+_41340520 | 0.20 |
ENSDART00000005127
|
garem
|
GRB2 associated, regulator of MAPK1 |
| chr22_+_20720808 | 0.19 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr23_-_524780 | 0.19 |
ENSDART00000055139
|
col9a3
|
collagen, type IX, alpha 3 |
| chr22_-_16180849 | 0.18 |
ENSDART00000090390
|
vcam1b
|
vascular cell adhesion molecule 1b |
| chr17_+_15297398 | 0.17 |
ENSDART00000156574
|
si:ch211-270g19.5
|
si:ch211-270g19.5 |
| chr19_+_390298 | 0.17 |
ENSDART00000136361
|
sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr20_+_6659770 | 0.16 |
ENSDART00000192135
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr5_-_2689753 | 0.16 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr19_-_11237125 | 0.16 |
ENSDART00000163921
|
ssr2
|
signal sequence receptor, beta |
| chr8_-_52859301 | 0.15 |
ENSDART00000162004
|
nr5a1a
|
nuclear receptor subfamily 5, group A, member 1a |
| chr3_+_29469283 | 0.15 |
ENSDART00000103592
|
fam83fa
|
family with sequence similarity 83, member Fa |
| chr19_+_5480327 | 0.14 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr22_-_10591876 | 0.14 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| chr2_+_38924975 | 0.13 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr5_-_43819663 | 0.13 |
ENSDART00000022481
|
mccc2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr14_-_21218891 | 0.12 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr24_-_24146875 | 0.11 |
ENSDART00000173052
|
map7d2b
|
MAP7 domain containing 2b |
| chr22_-_20720427 | 0.11 |
ENSDART00000105532
|
oaz1a
|
ornithine decarboxylase antizyme 1a |
| chr24_+_26895748 | 0.11 |
ENSDART00000089351
|
nceh1b.1
|
neutral cholesterol ester hydrolase 1b, tandem duplicate 1 |
| chr21_+_33172526 | 0.11 |
ENSDART00000183532
|
arl3l1
|
ADP-ribosylation factor-like 3, like 1 |
| chr23_+_44049509 | 0.11 |
ENSDART00000102003
|
txk
|
TXK tyrosine kinase |
| chr1_-_411331 | 0.10 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr14_+_36220479 | 0.10 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr4_+_70556298 | 0.10 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr10_+_19554604 | 0.09 |
ENSDART00000063806
|
atp6v1b2
|
ATPase H+ transporting V1 subunit B2 |
| chr20_+_27427112 | 0.09 |
ENSDART00000153459
|
si:dkey-22l1.1
|
si:dkey-22l1.1 |
| chr5_+_51443009 | 0.09 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr3_+_24537023 | 0.09 |
ENSDART00000077702
|
sp100.1
|
SP110 nuclear body protein, tandem duplicate 1 |
| chr13_+_35925490 | 0.09 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr6_+_40661703 | 0.07 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr13_-_31397987 | 0.07 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr4_+_332709 | 0.07 |
ENSDART00000067486
|
tulp4a
|
tubby like protein 4a |
| chr23_+_41800052 | 0.06 |
ENSDART00000141484
|
pdyn
|
prodynorphin |
| chr10_-_15919839 | 0.06 |
ENSDART00000065032
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr19_+_31061718 | 0.05 |
ENSDART00000145971
|
sostdc1b
|
sclerostin domain containing 1b |
| chr20_+_9211237 | 0.05 |
ENSDART00000139527
|
si:ch211-59d15.4
|
si:ch211-59d15.4 |
| chr5_-_49012569 | 0.05 |
ENSDART00000184690
|
si:dkey-172m14.2
|
si:dkey-172m14.2 |
| chr1_-_47431453 | 0.05 |
ENSDART00000101104
|
gja5b
|
gap junction protein, alpha 5b |
| chr22_-_34872533 | 0.05 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr25_+_13406069 | 0.05 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr23_+_40460333 | 0.05 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr11_+_14049131 | 0.04 |
ENSDART00000172471
|
grin3bb
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3Bb |
| chr22_-_9839003 | 0.04 |
ENSDART00000193573
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr12_+_499881 | 0.04 |
ENSDART00000167527
|
mprip
|
myosin phosphatase Rho interacting protein |
| chr23_+_9067131 | 0.04 |
ENSDART00000144533
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr21_-_26114886 | 0.04 |
ENSDART00000139320
|
nipal4
|
NIPA-like domain containing 4 |
| chr17_+_48164536 | 0.04 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr17_+_20639180 | 0.03 |
ENSDART00000063624
|
npy4r
|
neuropeptide Y receptor Y4 |
| chr3_+_23737795 | 0.03 |
ENSDART00000182247
|
hoxb3a
|
homeobox B3a |
| chr7_-_2039060 | 0.03 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr16_+_19029297 | 0.03 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr23_+_41799748 | 0.03 |
ENSDART00000144257
|
pdyn
|
prodynorphin |
| chr19_+_164013 | 0.03 |
ENSDART00000167379
|
carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr13_-_9886579 | 0.03 |
ENSDART00000101926
|
si:ch211-117n7.7
|
si:ch211-117n7.7 |
| chr5_-_21030934 | 0.03 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr14_+_40641350 | 0.02 |
ENSDART00000074506
|
tnmd
|
tenomodulin |
| chr15_+_9294620 | 0.02 |
ENSDART00000133588
ENSDART00000140009 |
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr5_+_42136359 | 0.02 |
ENSDART00000083731
|
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr8_+_22438398 | 0.02 |
ENSDART00000135145
|
zgc:153759
|
zgc:153759 |
| chr3_-_7106678 | 0.02 |
ENSDART00000175293
ENSDART00000187732 ENSDART00000158652 |
zgc:113102
BX005085.2
|
zgc:113102 |
| chr22_+_6757741 | 0.02 |
ENSDART00000140815
|
CT583625.1
|
|
| chr9_-_30363770 | 0.02 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr23_+_16910071 | 0.02 |
ENSDART00000104793
|
si:dkey-147f3.8
|
si:dkey-147f3.8 |
| chr5_+_61941610 | 0.02 |
ENSDART00000168808
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr17_+_27456804 | 0.01 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr22_-_14309817 | 0.01 |
ENSDART00000171625
|
si:ch73-338d8.5
|
si:ch73-338d8.5 |
| chr25_-_36827491 | 0.01 |
ENSDART00000170941
|
CU929365.1
|
|
| chr7_-_41014773 | 0.01 |
ENSDART00000013785
|
insig1
|
insulin induced gene 1 |
| chr5_-_14390445 | 0.01 |
ENSDART00000026120
|
ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr4_+_30785713 | 0.01 |
ENSDART00000165945
|
si:dkey-178j11.5
|
si:dkey-178j11.5 |
| chr5_+_64739762 | 0.01 |
ENSDART00000161112
ENSDART00000135610 ENSDART00000002908 |
olfm1a
|
olfactomedin 1a |
| chr2_+_47906240 | 0.01 |
ENSDART00000122206
|
ftr23
|
finTRIM family, member 23 |
| chr19_-_1920613 | 0.01 |
ENSDART00000186176
|
si:ch211-149a19.3
|
si:ch211-149a19.3 |
| chr20_-_4407228 | 0.01 |
ENSDART00000166997
|
CABZ01032982.1
|
|
| chr2_+_51194657 | 0.00 |
ENSDART00000161381
|
lrrc24
|
leucine rich repeat containing 24 |
| chr17_-_15657029 | 0.00 |
ENSDART00000153925
|
fut9a
|
fucosyltransferase 9a |
| chr22_-_6991514 | 0.00 |
ENSDART00000148233
|
fgfr1bl
|
fibroblast growth factor receptor 1b, like |
| chr23_+_804119 | 0.00 |
ENSDART00000148163
|
frmd4bb
|
FERM domain containing 4Bb |
| chr4_+_28997595 | 0.00 |
ENSDART00000133357
|
si:dkey-13e3.1
|
si:dkey-13e3.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of egr4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.5 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.3 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.1 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.0 | 0.1 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.0 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |