Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
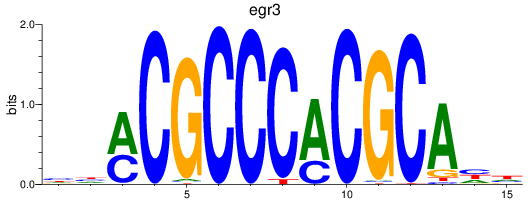
Results for egr3
Z-value: 0.35
Transcription factors associated with egr3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr3
|
ENSDARG00000089156 | early growth response 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr3 | dr11_v1_chr8_+_50727220_50727220 | -0.11 | 6.7e-01 | Click! |
Activity profile of egr3 motif
Sorted Z-values of egr3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_2584423 | 1.35 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr22_-_10591876 | 0.60 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| chr2_+_59015878 | 0.56 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr17_+_46387086 | 0.55 |
ENSDART00000157079
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr16_-_47381519 | 0.54 |
ENSDART00000032188
ENSDART00000150136 |
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr2_+_16710889 | 0.51 |
ENSDART00000017852
|
ubxn7
|
UBX domain protein 7 |
| chr7_+_46019780 | 0.51 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr12_-_990149 | 0.49 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr13_+_35925490 | 0.44 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr1_+_12295142 | 0.43 |
ENSDART00000158595
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr23_-_31645760 | 0.38 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr7_-_46019756 | 0.33 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr7_-_6364168 | 0.28 |
ENSDART00000173332
|
zgc:112234
|
zgc:112234 |
| chr2_+_105748 | 0.27 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr11_+_1565806 | 0.27 |
ENSDART00000137479
|
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr19_-_48039400 | 0.26 |
ENSDART00000166748
ENSDART00000165921 |
csf3b
|
colony stimulating factor 3 (granulocyte) b |
| chr13_-_36141486 | 0.24 |
ENSDART00000044115
|
med6
|
mediator complex subunit 6 |
| chr24_+_3328354 | 0.23 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr20_+_28803642 | 0.23 |
ENSDART00000188526
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr5_-_21030934 | 0.23 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr7_+_55518519 | 0.23 |
ENSDART00000098476
ENSDART00000149915 |
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr2_+_38924975 | 0.21 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr11_+_14049131 | 0.20 |
ENSDART00000172471
|
grin3bb
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3Bb |
| chr12_-_26537145 | 0.17 |
ENSDART00000138437
ENSDART00000163931 ENSDART00000132737 |
acsf2
|
acyl-CoA synthetase family member 2 |
| chr12_-_37449396 | 0.16 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr23_-_29556844 | 0.15 |
ENSDART00000138021
|
rbp7a
|
retinol binding protein 7a, cellular |
| chr8_+_6576940 | 0.15 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr5_+_29831235 | 0.14 |
ENSDART00000109660
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr4_-_73587205 | 0.14 |
ENSDART00000150837
|
znf1015
|
zinc finger protein 1015 |
| chr3_-_11624694 | 0.14 |
ENSDART00000127157
|
hlfa
|
hepatic leukemia factor a |
| chr16_-_12512568 | 0.13 |
ENSDART00000055161
ENSDART00000160906 |
cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr8_-_4618653 | 0.13 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr5_+_6617401 | 0.11 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr20_+_27712714 | 0.11 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr2_+_35880600 | 0.11 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr19_+_5480327 | 0.10 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr3_+_22984098 | 0.10 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr18_+_3169579 | 0.10 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr15_+_9294620 | 0.09 |
ENSDART00000133588
ENSDART00000140009 |
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr21_-_217589 | 0.09 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr3_-_7106678 | 0.08 |
ENSDART00000175293
ENSDART00000187732 ENSDART00000158652 |
zgc:113102
BX005085.2
|
zgc:113102 |
| chr4_+_63484571 | 0.07 |
ENSDART00000169518
ENSDART00000168681 |
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr2_+_23062085 | 0.07 |
ENSDART00000153745
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr13_-_49666615 | 0.06 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr24_-_39354829 | 0.06 |
ENSDART00000169108
|
kcnj12a
|
potassium inwardly-rectifying channel, subfamily J, member 12a |
| chr3_-_49504023 | 0.06 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr13_-_31166544 | 0.05 |
ENSDART00000146250
ENSDART00000132129 ENSDART00000139591 |
mapk8a
|
mitogen-activated protein kinase 8a |
| chr9_-_30363770 | 0.05 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr1_-_48922273 | 0.05 |
ENSDART00000150254
|
si:ch211-112b1.2
|
si:ch211-112b1.2 |
| chr23_+_804119 | 0.04 |
ENSDART00000148163
|
frmd4bb
|
FERM domain containing 4Bb |
| chr15_-_14038227 | 0.04 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr11_-_2594045 | 0.04 |
ENSDART00000114079
|
nab2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr4_+_30785713 | 0.03 |
ENSDART00000165945
|
si:dkey-178j11.5
|
si:dkey-178j11.5 |
| chr17_+_48164536 | 0.02 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr15_-_16121496 | 0.02 |
ENSDART00000128624
|
sgk494a
|
uncharacterized serine/threonine-protein kinase SgK494a |
| chr21_-_24865217 | 0.02 |
ENSDART00000101136
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr22_+_9806867 | 0.02 |
ENSDART00000137182
ENSDART00000106103 |
si:ch211-236g6.1
|
si:ch211-236g6.1 |
| chr20_+_13894123 | 0.01 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr22_+_1806235 | 0.01 |
ENSDART00000158966
|
znf1168
|
zinc finger protein 1168 |
| chr21_-_24865454 | 0.01 |
ENSDART00000142907
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr3_-_38918697 | 0.01 |
ENSDART00000145630
|
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr9_-_48036 | 0.01 |
ENSDART00000165230
ENSDART00000171335 |
map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr12_-_44016898 | 0.00 |
ENSDART00000175304
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of egr3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.4 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 1.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.4 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.2 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.5 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.4 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.5 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.2 | GO:0031956 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |