Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
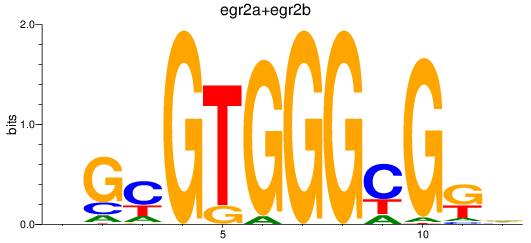
Results for egr2a+egr2b
Z-value: 0.99
Transcription factors associated with egr2a+egr2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr2b
|
ENSDARG00000042826 | early growth response 2b |
|
egr2a
|
ENSDARG00000044098 | early growth response 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr2a | dr11_v1_chr17_-_43666166_43666166 | 0.97 | 1.8e-11 | Click! |
| egr2b | dr11_v1_chr12_-_8486330_8486330 | 0.45 | 6.1e-02 | Click! |
Activity profile of egr2a+egr2b motif
Sorted Z-values of egr2a+egr2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_10560964 | 4.58 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr16_+_10963602 | 3.40 |
ENSDART00000141032
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr16_-_54405976 | 3.20 |
ENSDART00000055395
|
osr2
|
odd-skipped related transciption factor 2 |
| chr16_+_23978978 | 2.99 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr15_-_20939579 | 2.98 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr23_+_39606108 | 2.82 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr14_+_80685 | 2.68 |
ENSDART00000188443
|
stag3
|
stromal antigen 3 |
| chr20_-_22778394 | 2.61 |
ENSDART00000152645
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr3_+_60007703 | 2.61 |
ENSDART00000157351
ENSDART00000153928 ENSDART00000155876 |
si:ch211-110e21.3
|
si:ch211-110e21.3 |
| chr15_+_31526225 | 2.31 |
ENSDART00000154456
|
wdr95
|
WD40 repeat domain 95 |
| chr18_+_402048 | 2.30 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr15_-_20916251 | 2.26 |
ENSDART00000134053
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr12_+_49125510 | 2.18 |
ENSDART00000185804
|
FO704607.1
|
|
| chr25_+_24250247 | 2.10 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr16_+_10918252 | 2.08 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr5_-_26093945 | 2.06 |
ENSDART00000010199
ENSDART00000145096 |
fam219ab
|
family with sequence similarity 219, member Ab |
| chr21_-_308852 | 1.91 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr5_-_72136548 | 1.89 |
ENSDART00000007827
|
spra
|
sepiapterin reductase a |
| chr3_-_10779777 | 1.87 |
ENSDART00000153911
|
znf1004
|
zinc finger protein 1004 |
| chr20_+_27242595 | 1.85 |
ENSDART00000149300
|
si:dkey-85n7.6
|
si:dkey-85n7.6 |
| chr14_-_2933185 | 1.82 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr17_-_52643970 | 1.77 |
ENSDART00000190594
|
spred1
|
sprouty-related, EVH1 domain containing 1 |
| chr23_+_20422661 | 1.77 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr9_+_17348745 | 1.76 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr7_-_22981796 | 1.73 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr6_+_26314464 | 1.70 |
ENSDART00000115392
|
dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr3_+_32492467 | 1.67 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr24_+_3963684 | 1.66 |
ENSDART00000182959
ENSDART00000185926 ENSDART00000167043 ENSDART00000033394 |
pfkpa
|
phosphofructokinase, platelet a |
| chr10_-_36726188 | 1.66 |
ENSDART00000157262
|
pimr137
|
Pim proto-oncogene, serine/threonine kinase, related 137 |
| chr6_-_35446110 | 1.59 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr7_-_33023404 | 1.58 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr19_-_31341850 | 1.48 |
ENSDART00000040810
|
arl4ab
|
ADP-ribosylation factor-like 4ab |
| chr14_-_15171435 | 1.48 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr20_-_51697437 | 1.46 |
ENSDART00000145391
|
si:ch211-14a11.2
|
si:ch211-14a11.2 |
| chr22_-_34872533 | 1.46 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr10_+_45345574 | 1.46 |
ENSDART00000166085
|
ppiab
|
peptidylprolyl isomerase Ab (cyclophilin A) |
| chr5_-_68022631 | 1.46 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr16_+_11029762 | 1.41 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr3_+_40809011 | 1.40 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr16_+_51180938 | 1.40 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr3_+_40315410 | 1.39 |
ENSDART00000083241
ENSDART00000132827 |
slc29a4
|
solute carrier family 29 (equilibrative nucleoside transporter), member 4 |
| chr3_+_14388010 | 1.36 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr2_+_26288301 | 1.36 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr2_-_17827983 | 1.35 |
ENSDART00000166518
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr13_-_37653840 | 1.33 |
ENSDART00000143806
|
si:dkey-188i13.11
|
si:dkey-188i13.11 |
| chr24_+_26997798 | 1.33 |
ENSDART00000089506
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr12_+_695619 | 1.33 |
ENSDART00000161691
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr5_-_2689753 | 1.33 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr17_-_6599484 | 1.33 |
ENSDART00000156927
|
ANKRD66
|
si:ch211-189e2.2 |
| chr10_+_10351685 | 1.31 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr22_-_367569 | 1.31 |
ENSDART00000041895
|
ssu72
|
SSU72 homolog, RNA polymerase II CTD phosphatase |
| chr16_+_40575742 | 1.30 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr8_+_8298439 | 1.29 |
ENSDART00000170566
|
srpk3
|
SRSF protein kinase 3 |
| chr19_+_636886 | 1.28 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr1_-_411331 | 1.28 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr3_-_32180796 | 1.27 |
ENSDART00000133191
ENSDART00000055308 |
pih1d1
|
PIH1 domain containing 1 |
| chr25_+_37348730 | 1.26 |
ENSDART00000156639
|
pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr2_-_17828279 | 1.24 |
ENSDART00000083314
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr14_-_36863432 | 1.23 |
ENSDART00000158052
|
rnf130
|
ring finger protein 130 |
| chr18_+_33788340 | 1.23 |
ENSDART00000136950
|
si:dkey-145c18.2
|
si:dkey-145c18.2 |
| chr20_+_1398564 | 1.23 |
ENSDART00000002242
|
leg1.2
|
liver-enriched gene 1, tandem duplicate 2 |
| chr12_+_13282797 | 1.23 |
ENSDART00000137757
ENSDART00000152397 |
irf9
|
interferon regulatory factor 9 |
| chr12_-_212843 | 1.22 |
ENSDART00000083574
|
CABZ01102039.1
|
|
| chr23_+_9560991 | 1.21 |
ENSDART00000081433
ENSDART00000131594 ENSDART00000130069 ENSDART00000138601 |
adrm1
|
adhesion regulating molecule 1 |
| chr7_+_38510197 | 1.21 |
ENSDART00000173468
ENSDART00000100479 |
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr3_+_1211242 | 1.20 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr20_+_27298783 | 1.19 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr22_-_16180849 | 1.19 |
ENSDART00000090390
|
vcam1b
|
vascular cell adhesion molecule 1b |
| chr23_+_9560797 | 1.19 |
ENSDART00000180014
|
adrm1
|
adhesion regulating molecule 1 |
| chr16_-_21668082 | 1.15 |
ENSDART00000088513
|
gnl1
|
guanine nucleotide binding protein-like 1 |
| chr5_-_12093618 | 1.15 |
ENSDART00000161542
|
lrrc74b
|
leucine rich repeat containing 74B |
| chr5_-_69523816 | 1.14 |
ENSDART00000112692
|
si:ch211-157p22.10
|
si:ch211-157p22.10 |
| chr8_+_14158021 | 1.12 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr7_+_20563305 | 1.10 |
ENSDART00000169661
|
si:dkey-19b23.10
|
si:dkey-19b23.10 |
| chr15_+_3284684 | 1.09 |
ENSDART00000179778
|
foxo1a
|
forkhead box O1 a |
| chr16_-_12173554 | 1.08 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr13_-_438705 | 1.07 |
ENSDART00000082142
|
CU570800.1
|
|
| chr8_-_52859301 | 1.07 |
ENSDART00000162004
|
nr5a1a
|
nuclear receptor subfamily 5, group A, member 1a |
| chr4_+_1600034 | 1.06 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr25_-_16146851 | 1.06 |
ENSDART00000104043
|
dkk3b
|
dickkopf WNT signaling pathway inhibitor 3b |
| chr20_-_45661049 | 1.05 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr10_+_15777064 | 1.05 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr6_+_21740672 | 1.04 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr19_+_935565 | 1.03 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr21_+_11415224 | 1.02 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr6_+_2093367 | 1.02 |
ENSDART00000148396
|
tgm2b
|
transglutaminase 2b |
| chr20_+_13894123 | 1.01 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr11_-_4302251 | 1.00 |
ENSDART00000182554
|
CT033790.1
|
|
| chr10_+_40583930 | 0.98 |
ENSDART00000134523
|
taar18j
|
trace amine associated receptor 18j |
| chr23_+_44307996 | 0.98 |
ENSDART00000042430
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr11_+_1796426 | 0.98 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr5_+_483965 | 0.97 |
ENSDART00000150007
|
tek
|
TEK tyrosine kinase, endothelial |
| chr13_+_51853716 | 0.97 |
ENSDART00000175341
ENSDART00000187855 |
LT631684.1
|
|
| chr11_+_30161168 | 0.97 |
ENSDART00000157385
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr16_-_12512568 | 0.97 |
ENSDART00000055161
ENSDART00000160906 |
cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr5_-_46273938 | 0.97 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr13_-_18069421 | 0.97 |
ENSDART00000146772
ENSDART00000134477 |
zfand4
|
zinc finger, AN1-type domain 4 |
| chr12_-_20303438 | 0.95 |
ENSDART00000153057
|
rhbdf1b
|
rhomboid 5 homolog 1b (Drosophila) |
| chr12_+_2522642 | 0.95 |
ENSDART00000152567
|
frmpd2
|
FERM and PDZ domain containing 2 |
| chr8_-_15292197 | 0.94 |
ENSDART00000140867
|
spata6
|
spermatogenesis associated 6 |
| chr15_+_3284416 | 0.94 |
ENSDART00000187665
ENSDART00000171723 |
foxo1a
|
forkhead box O1 a |
| chr14_+_29769336 | 0.93 |
ENSDART00000105898
|
si:dkey-34l15.1
|
si:dkey-34l15.1 |
| chr25_+_34013093 | 0.93 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr18_+_30028135 | 0.93 |
ENSDART00000140825
ENSDART00000145306 ENSDART00000136810 |
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr9_+_3035128 | 0.93 |
ENSDART00000140693
|
si:ch211-173m16.2
|
si:ch211-173m16.2 |
| chr12_+_499881 | 0.92 |
ENSDART00000167527
|
mprip
|
myosin phosphatase Rho interacting protein |
| chr25_+_34915576 | 0.90 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr16_-_12173399 | 0.89 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr4_+_16710001 | 0.89 |
ENSDART00000035899
|
pkp2
|
plakophilin 2 |
| chr17_+_53311618 | 0.87 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr15_+_42397125 | 0.86 |
ENSDART00000169751
|
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr5_-_52784152 | 0.85 |
ENSDART00000169307
|
fam189a2
|
family with sequence similarity 189, member A2 |
| chr25_-_23526058 | 0.85 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr1_-_59417949 | 0.85 |
ENSDART00000170558
|
si:ch211-188p14.3
|
si:ch211-188p14.3 |
| chr24_+_42131564 | 0.84 |
ENSDART00000153854
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr24_+_42127983 | 0.83 |
ENSDART00000190157
ENSDART00000176032 ENSDART00000175790 |
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr7_+_41340520 | 0.83 |
ENSDART00000005127
|
garem
|
GRB2 associated, regulator of MAPK1 |
| chr20_-_54381034 | 0.83 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr1_-_59176949 | 0.82 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr20_-_5369105 | 0.82 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr1_+_55293424 | 0.82 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr6_-_15604157 | 0.81 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr10_-_27196093 | 0.81 |
ENSDART00000185282
|
auts2a
|
autism susceptibility candidate 2a |
| chr13_+_36633355 | 0.81 |
ENSDART00000135612
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr3_+_24537023 | 0.81 |
ENSDART00000077702
|
sp100.1
|
SP110 nuclear body protein, tandem duplicate 1 |
| chr14_+_15495088 | 0.80 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr21_-_7882905 | 0.80 |
ENSDART00000056561
|
s100z
|
S100 calcium binding protein Z |
| chr11_+_3254524 | 0.80 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr25_+_34915762 | 0.80 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr21_+_103194 | 0.80 |
ENSDART00000162755
|
zer1
|
zyg-11 related, cell cycle regulator |
| chr13_+_25486608 | 0.79 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr11_-_236766 | 0.79 |
ENSDART00000163978
|
dusp7
|
dual specificity phosphatase 7 |
| chr8_+_53051701 | 0.78 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr22_-_16180467 | 0.78 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr11_+_40306579 | 0.77 |
ENSDART00000167504
|
si:dkey-207b20.2
|
si:dkey-207b20.2 |
| chr11_+_3254252 | 0.76 |
ENSDART00000123568
|
pmela
|
premelanosome protein a |
| chr1_-_46632948 | 0.74 |
ENSDART00000148893
ENSDART00000053232 |
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr4_-_9852318 | 0.74 |
ENSDART00000080702
|
glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr18_-_3166726 | 0.74 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr15_-_23793641 | 0.74 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr22_+_4707663 | 0.74 |
ENSDART00000042194
|
cers4a
|
ceramide synthase 4a |
| chr23_+_44374041 | 0.73 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr12_+_18782821 | 0.73 |
ENSDART00000152918
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr18_+_5490668 | 0.73 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr3_+_36284986 | 0.73 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr14_+_48960078 | 0.73 |
ENSDART00000165514
|
mif
|
macrophage migration inhibitory factor |
| chr14_+_15257658 | 0.73 |
ENSDART00000161625
ENSDART00000193577 |
si:dkey-77g12.4
si:dkey-203a12.5
|
si:dkey-77g12.4 si:dkey-203a12.5 |
| chr18_+_16246806 | 0.72 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr7_-_7845540 | 0.72 |
ENSDART00000166280
|
cxcl8b.1
|
chemokine (C-X-C motif) ligand 8b, duplicate 1 |
| chr10_+_6884123 | 0.71 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr7_+_25053331 | 0.71 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr2_+_21486529 | 0.71 |
ENSDART00000047468
|
inhbab
|
inhibin, beta Ab |
| chr11_-_236984 | 0.71 |
ENSDART00000170778
|
dusp7
|
dual specificity phosphatase 7 |
| chr9_-_23891102 | 0.70 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr9_-_9671244 | 0.70 |
ENSDART00000018228
|
gsk3b
|
glycogen synthase kinase 3 beta |
| chr3_-_27601544 | 0.68 |
ENSDART00000188201
|
zgc:66160
|
zgc:66160 |
| chr22_-_50878 | 0.67 |
ENSDART00000166132
|
CABZ01085140.1
|
|
| chr5_+_22969651 | 0.67 |
ENSDART00000089992
ENSDART00000145477 |
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr13_+_33055548 | 0.66 |
ENSDART00000137315
|
rdh12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis) |
| chr8_-_53490376 | 0.66 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr14_+_30910114 | 0.66 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr22_+_6905210 | 0.66 |
ENSDART00000167530
|
CABZ01065328.2
|
|
| chr14_-_21218891 | 0.66 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr5_+_51443009 | 0.66 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr8_+_2456854 | 0.66 |
ENSDART00000133938
ENSDART00000002764 |
polb
|
polymerase (DNA directed), beta |
| chr25_-_8602437 | 0.65 |
ENSDART00000171200
|
rhcgb
|
Rh family, C glycoprotein b |
| chrM_+_9735 | 0.65 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr16_+_28754403 | 0.64 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr3_-_30061985 | 0.64 |
ENSDART00000189583
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr20_-_53366137 | 0.64 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr20_+_16173618 | 0.64 |
ENSDART00000192109
ENSDART00000104112 ENSDART00000129633 |
zyg11
|
zyg-11 homolog (C. elegans) |
| chr11_+_41560792 | 0.63 |
ENSDART00000127292
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr6_+_40661703 | 0.62 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr1_-_44940830 | 0.62 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr1_-_59571758 | 0.61 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr5_+_36513605 | 0.61 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr1_-_19845378 | 0.61 |
ENSDART00000139314
ENSDART00000132958 ENSDART00000147502 |
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr17_+_30587333 | 0.61 |
ENSDART00000156500
|
nhsl1a
|
NHS-like 1a |
| chr4_+_70556298 | 0.58 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr21_+_7900107 | 0.58 |
ENSDART00000056560
|
ch25hl2
|
cholesterol 25-hydroxylase like 2 |
| chr3_+_62217479 | 0.58 |
ENSDART00000175012
|
BX470259.4
|
|
| chr6_-_15604417 | 0.58 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr4_+_39742119 | 0.57 |
ENSDART00000176004
|
CR749167.2
|
|
| chr2_-_48966431 | 0.57 |
ENSDART00000147948
|
kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr8_+_23436411 | 0.56 |
ENSDART00000140044
|
foxp3b
|
forkhead box P3b |
| chr9_-_2573121 | 0.56 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr15_+_1397811 | 0.56 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr24_+_35564668 | 0.56 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr4_-_12914163 | 0.55 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr7_+_26173751 | 0.55 |
ENSDART00000065131
|
si:ch211-196f2.7
|
si:ch211-196f2.7 |
| chr19_-_617246 | 0.55 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr14_+_36220479 | 0.54 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr24_+_37406535 | 0.54 |
ENSDART00000138264
|
si:ch211-183d21.1
|
si:ch211-183d21.1 |
| chr5_-_67911111 | 0.54 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr18_+_33762090 | 0.54 |
ENSDART00000147605
|
si:dkey-145c18.4
|
si:dkey-145c18.4 |
| chr9_-_8454060 | 0.54 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr7_+_7048245 | 0.53 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr11_-_44347587 | 0.53 |
ENSDART00000172919
|
si:cabz01100188.1
|
si:cabz01100188.1 |
| chr3_-_27601812 | 0.53 |
ENSDART00000077734
|
zgc:66160
|
zgc:66160 |
| chr3_-_61185746 | 0.53 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of egr2a+egr2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.6 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.6 | 1.7 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.5 | 1.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.4 | 1.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.4 | 1.3 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.4 | 1.1 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.3 | 1.4 | GO:0060468 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.3 | 3.0 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 1.0 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.3 | 0.9 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.3 | 1.8 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.3 | 2.0 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.3 | 2.6 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.3 | 1.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.3 | 0.8 | GO:0010660 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.3 | 1.0 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.2 | 1.9 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) pteridine-containing compound biosynthetic process(GO:0042559) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 0.7 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 2.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 1.9 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.2 | 1.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 0.5 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 2.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 1.7 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.7 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.8 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 2.1 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 2.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.6 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.1 | 0.9 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.9 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.5 | GO:0021828 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.1 | 0.5 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.5 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 1.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 0.6 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 1.4 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 2.3 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.1 | 1.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.3 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.1 | 0.5 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.5 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.3 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 1.1 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 0.1 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.1 | 0.8 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.1 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.1 | 1.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 0.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.4 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 1.0 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.1 | GO:0021572 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) |
| 0.1 | 0.6 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.2 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 1.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.7 | GO:0008406 | gonad development(GO:0008406) |
| 0.0 | 0.3 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 1.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.5 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.0 | 3.5 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 1.5 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.0 | 0.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.2 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 0.4 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.9 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.8 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 2.4 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.4 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.3 | GO:0042664 | negative regulation of endodermal cell fate specification(GO:0042664) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.5 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 4.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.5 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.2 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 1.6 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.7 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 1.1 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.5 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.7 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 1.7 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.9 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.7 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.9 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.2 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.7 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.0 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 1.7 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.5 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.2 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.1 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.4 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.3 | 1.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 1.3 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.3 | 1.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 1.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.3 | 4.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.3 | 1.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 2.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 0.9 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.2 | 1.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 2.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.0 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.6 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 1.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 1.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 1.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.7 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.5 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.1 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.4 | 2.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 1.3 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.3 | 0.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.3 | 2.2 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.2 | 2.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 1.0 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 0.6 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.7 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.2 | 1.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 1.4 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.2 | 2.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 1.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.4 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 3.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.7 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 2.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.4 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 1.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.6 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 1.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.4 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 1.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 4.6 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.1 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.3 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 1.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.6 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 4.5 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.7 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 3.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.3 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.1 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.3 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 2.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 1.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 5.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 1.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 1.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.9 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.6 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.0 | 0.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |