Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
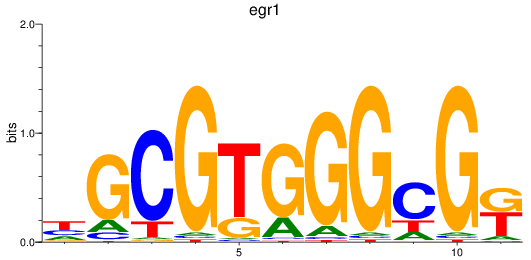
Results for egr1
Z-value: 0.48
Transcription factors associated with egr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr1
|
ENSDARG00000037421 | early growth response 1 |
|
egr1
|
ENSDARG00000115563 | early growth response 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr1 | dr11_v1_chr14_-_21336385_21336385 | 0.45 | 6.3e-02 | Click! |
Activity profile of egr1 motif
Sorted Z-values of egr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_54356272 | 0.98 |
ENSDART00000145735
|
znf410
|
zinc finger protein 410 |
| chr23_+_39606108 | 0.89 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr16_-_54405976 | 0.86 |
ENSDART00000055395
|
osr2
|
odd-skipped related transciption factor 2 |
| chr2_-_22535 | 0.72 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr14_+_48960078 | 0.70 |
ENSDART00000165514
|
mif
|
macrophage migration inhibitory factor |
| chr20_-_51697437 | 0.68 |
ENSDART00000145391
|
si:ch211-14a11.2
|
si:ch211-14a11.2 |
| chr20_-_52631998 | 0.66 |
ENSDART00000145283
ENSDART00000139072 |
si:ch211-221n20.1
|
si:ch211-221n20.1 |
| chr3_-_32180796 | 0.63 |
ENSDART00000133191
ENSDART00000055308 |
pih1d1
|
PIH1 domain containing 1 |
| chr19_-_25427255 | 0.63 |
ENSDART00000036854
|
glcci1a
|
glucocorticoid induced 1a |
| chr16_-_12512568 | 0.61 |
ENSDART00000055161
ENSDART00000160906 |
cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr1_-_8653385 | 0.61 |
ENSDART00000193041
|
actb1
|
actin, beta 1 |
| chr14_+_2243 | 0.61 |
ENSDART00000191193
|
CYTL1
|
cytokine like 1 |
| chr12_-_990149 | 0.60 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr20_-_22778394 | 0.57 |
ENSDART00000152645
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr24_+_24923166 | 0.57 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr2_+_16710889 | 0.56 |
ENSDART00000017852
|
ubxn7
|
UBX domain protein 7 |
| chr7_-_10560964 | 0.55 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr18_+_5490668 | 0.54 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr19_+_7154500 | 0.54 |
ENSDART00000035967
ENSDART00000160894 |
brd2a
|
bromodomain containing 2a |
| chr5_+_337215 | 0.53 |
ENSDART00000167982
|
rnf170
|
ring finger protein 170 |
| chr17_+_24821627 | 0.53 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr17_+_53311618 | 0.53 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr21_-_308852 | 0.52 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr22_-_80533 | 0.52 |
ENSDART00000183124
|
CABZ01101813.1
|
|
| chr1_-_49434798 | 0.51 |
ENSDART00000150842
ENSDART00000150789 |
ATP5MD
|
si:dkeyp-80c12.10 |
| chr20_+_13894123 | 0.49 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr24_+_3328354 | 0.49 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr16_-_13595027 | 0.49 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr15_-_20939579 | 0.49 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr16_+_23978978 | 0.47 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr19_+_48102560 | 0.45 |
ENSDART00000164464
|
utp18
|
UTP18 small subunit (SSU) processome component |
| chr17_-_2584423 | 0.45 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr14_+_17397534 | 0.44 |
ENSDART00000180162
ENSDART00000129838 |
rnf212
|
ring finger protein 212 |
| chr9_-_52386733 | 0.43 |
ENSDART00000171721
|
dap1b
|
death associated protein 1b |
| chr7_+_20563305 | 0.42 |
ENSDART00000169661
|
si:dkey-19b23.10
|
si:dkey-19b23.10 |
| chr2_-_48753873 | 0.41 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr18_-_50956280 | 0.41 |
ENSDART00000058457
|
st7
|
suppression of tumorigenicity 7 |
| chr18_+_36601556 | 0.41 |
ENSDART00000127825
|
clasrp
|
CLK4-associating serine/arginine rich protein |
| chr24_+_37406535 | 0.40 |
ENSDART00000138264
|
si:ch211-183d21.1
|
si:ch211-183d21.1 |
| chr25_-_16146851 | 0.40 |
ENSDART00000104043
|
dkk3b
|
dickkopf WNT signaling pathway inhibitor 3b |
| chr19_+_636886 | 0.38 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr5_+_72377851 | 0.38 |
ENSDART00000160479
|
lman2la
|
lectin, mannose-binding 2-like a |
| chr18_+_36601403 | 0.38 |
ENSDART00000192905
|
clasrp
|
CLK4-associating serine/arginine rich protein |
| chr2_+_105748 | 0.38 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr24_+_42127983 | 0.36 |
ENSDART00000190157
ENSDART00000176032 ENSDART00000175790 |
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr12_+_48851381 | 0.35 |
ENSDART00000187241
ENSDART00000187796 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr24_-_37688398 | 0.34 |
ENSDART00000141414
|
zgc:112185
|
zgc:112185 |
| chr6_-_35446110 | 0.33 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr21_-_280769 | 0.33 |
ENSDART00000157753
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr6_+_2093367 | 0.32 |
ENSDART00000148396
|
tgm2b
|
transglutaminase 2b |
| chr7_+_41340520 | 0.31 |
ENSDART00000005127
|
garem
|
GRB2 associated, regulator of MAPK1 |
| chr12_-_49168398 | 0.31 |
ENSDART00000186608
|
FO704624.1
|
|
| chr2_-_54639964 | 0.31 |
ENSDART00000100103
|
acss2l
|
acyl-CoA synthetase short chain family member 2 like |
| chr16_+_10918252 | 0.30 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr10_-_35030673 | 0.30 |
ENSDART00000135338
|
supt20
|
SPT20 homolog, SAGA complex component |
| chr15_+_3284684 | 0.29 |
ENSDART00000179778
|
foxo1a
|
forkhead box O1 a |
| chr16_+_4658250 | 0.29 |
ENSDART00000006212
|
LIN28A
|
si:ch1073-284b18.2 |
| chr7_-_46019756 | 0.28 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr5_+_36513605 | 0.27 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr9_-_24413008 | 0.27 |
ENSDART00000135897
|
tmeff2a
|
transmembrane protein with EGF-like and two follistatin-like domains 2a |
| chr3_-_22829710 | 0.26 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr20_-_45661049 | 0.26 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr18_+_402048 | 0.26 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr5_-_2689753 | 0.25 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr15_-_47848544 | 0.24 |
ENSDART00000098711
|
eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr8_-_52859301 | 0.24 |
ENSDART00000162004
|
nr5a1a
|
nuclear receptor subfamily 5, group A, member 1a |
| chr19_-_11237125 | 0.24 |
ENSDART00000163921
|
ssr2
|
signal sequence receptor, beta |
| chr23_+_44374041 | 0.23 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr14_-_15699528 | 0.23 |
ENSDART00000161123
|
neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr16_-_13965518 | 0.23 |
ENSDART00000138304
|
si:dkey-85k15.4
|
si:dkey-85k15.4 |
| chr21_+_103194 | 0.23 |
ENSDART00000162755
|
zer1
|
zyg-11 related, cell cycle regulator |
| chr18_-_50845804 | 0.22 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr25_-_37501371 | 0.22 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr2_-_17827983 | 0.22 |
ENSDART00000166518
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr6_+_21740672 | 0.21 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr15_+_3284416 | 0.21 |
ENSDART00000187665
ENSDART00000171723 |
foxo1a
|
forkhead box O1 a |
| chr11_+_45436703 | 0.21 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr11_+_45461419 | 0.20 |
ENSDART00000173424
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr13_+_12739283 | 0.20 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr17_+_53311243 | 0.20 |
ENSDART00000160241
ENSDART00000160009 ENSDART00000162239 |
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr7_-_22981796 | 0.20 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr1_-_8917902 | 0.19 |
ENSDART00000137900
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr3_+_40809011 | 0.19 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr18_-_5103931 | 0.19 |
ENSDART00000188091
|
pdcd10a
|
programmed cell death 10a |
| chr6_-_15604157 | 0.19 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr3_+_32492467 | 0.18 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr2_-_17828279 | 0.18 |
ENSDART00000083314
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr3_+_45687266 | 0.18 |
ENSDART00000131652
|
gpr146
|
G protein-coupled receptor 146 |
| chr21_-_22325124 | 0.18 |
ENSDART00000142100
|
gdpd4b
|
glycerophosphodiester phosphodiesterase domain containing 4b |
| chr8_-_34762163 | 0.18 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr3_-_61185746 | 0.18 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr10_+_252425 | 0.18 |
ENSDART00000059478
|
lrrc32
|
leucine rich repeat containing 32 |
| chr21_+_11415224 | 0.18 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr14_-_21218891 | 0.17 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr15_+_43906043 | 0.17 |
ENSDART00000010881
|
naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr1_-_411331 | 0.16 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr15_-_17099560 | 0.16 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr6_-_15604417 | 0.16 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr22_+_38192568 | 0.16 |
ENSDART00000126323
|
cp
|
ceruloplasmin |
| chr14_-_14566417 | 0.15 |
ENSDART00000159056
|
si:dkey-27i16.2
|
si:dkey-27i16.2 |
| chr12_+_41510492 | 0.15 |
ENSDART00000170976
ENSDART00000176164 |
kif5bb
|
kinesin family member 5B, b |
| chr20_-_26383368 | 0.15 |
ENSDART00000024518
ENSDART00000087844 |
esr1
|
estrogen receptor 1 |
| chr2_+_38924975 | 0.15 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr7_+_22981909 | 0.15 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr17_-_25831569 | 0.15 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr4_+_70556298 | 0.15 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr8_-_53490376 | 0.15 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr9_-_105135 | 0.15 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr6_+_59808677 | 0.14 |
ENSDART00000164469
|
caskb
|
calcium/calmodulin-dependent serine protein kinase b |
| chr12_-_15620090 | 0.14 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr21_+_3941758 | 0.14 |
ENSDART00000181345
|
setx
|
senataxin |
| chr3_-_40965596 | 0.14 |
ENSDART00000137188
|
cyp3c2
|
cytochrome P450, family 3, subfamily c, polypeptide 2 |
| chr12_-_20303438 | 0.14 |
ENSDART00000153057
|
rhbdf1b
|
rhomboid 5 homolog 1b (Drosophila) |
| chr19_+_935565 | 0.14 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr1_-_59571758 | 0.14 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr3_-_15444396 | 0.14 |
ENSDART00000104361
|
si:dkey-56d12.4
|
si:dkey-56d12.4 |
| chr5_+_43870389 | 0.13 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr2_-_48966431 | 0.13 |
ENSDART00000147948
|
kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr18_+_16246806 | 0.13 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr23_-_29556844 | 0.13 |
ENSDART00000138021
|
rbp7a
|
retinol binding protein 7a, cellular |
| chr14_+_36220479 | 0.13 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr25_-_32888115 | 0.13 |
ENSDART00000087586
|
c2cd4a
|
C2 calcium dependent domain containing 4A |
| chr25_+_37348730 | 0.13 |
ENSDART00000156639
|
pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr1_+_54673846 | 0.12 |
ENSDART00000145018
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr23_+_20422661 | 0.12 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr19_-_48039400 | 0.12 |
ENSDART00000166748
ENSDART00000165921 |
csf3b
|
colony stimulating factor 3 (granulocyte) b |
| chr20_-_54381034 | 0.12 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr21_-_33126697 | 0.12 |
ENSDART00000189293
ENSDART00000084559 |
CU019662.1
|
|
| chr2_+_20793982 | 0.12 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr19_+_58954 | 0.12 |
ENSDART00000162379
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr9_+_51784922 | 0.12 |
ENSDART00000125850
|
cd302
|
CD302 molecule |
| chr21_+_7900107 | 0.11 |
ENSDART00000056560
|
ch25hl2
|
cholesterol 25-hydroxylase like 2 |
| chr25_+_1304173 | 0.11 |
ENSDART00000155229
|
rxfp3.3b
|
relaxin/insulin-like family peptide receptor 3.3b |
| chr11_+_1565806 | 0.11 |
ENSDART00000137479
|
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr16_-_6821927 | 0.11 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr15_-_19250543 | 0.11 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr6_-_13206255 | 0.11 |
ENSDART00000065373
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr1_-_59422880 | 0.11 |
ENSDART00000167244
|
si:ch211-188p14.2
|
si:ch211-188p14.2 |
| chr22_+_1751640 | 0.11 |
ENSDART00000162093
|
znf1169
|
zinc finger protein 1169 |
| chr14_+_7939398 | 0.11 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr25_+_36315127 | 0.10 |
ENSDART00000191984
|
zgc:165555
|
zgc:165555 |
| chr15_-_47822597 | 0.10 |
ENSDART00000193236
ENSDART00000161391 |
CZQB01095947.1
|
|
| chr11_-_42418374 | 0.10 |
ENSDART00000160704
|
slmapa
|
sarcolemma associated protein a |
| chr18_-_49078428 | 0.10 |
ENSDART00000160702
ENSDART00000174103 |
BX663503.3
|
|
| chr4_-_23839789 | 0.10 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr10_+_2742499 | 0.10 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr5_+_15667874 | 0.10 |
ENSDART00000127015
|
srrm4
|
serine/arginine repetitive matrix 4 |
| chr10_-_15919839 | 0.10 |
ENSDART00000065032
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr4_-_64709908 | 0.09 |
ENSDART00000161032
|
si:dkey-9i5.2
|
si:dkey-9i5.2 |
| chr21_+_19517492 | 0.08 |
ENSDART00000123168
ENSDART00000187993 |
gzmk
|
granzyme K |
| chr6_+_14301214 | 0.08 |
ENSDART00000129491
|
tmem182b
|
transmembrane protein 182b |
| chr1_-_53259746 | 0.08 |
ENSDART00000134222
|
il15
|
interleukin 15 |
| chr4_+_39742119 | 0.08 |
ENSDART00000176004
|
CR749167.2
|
|
| chr11_+_37251825 | 0.08 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr14_+_32964 | 0.08 |
ENSDART00000166173
|
lyar
|
Ly1 antibody reactive homolog (mouse) |
| chr8_-_15292197 | 0.08 |
ENSDART00000140867
|
spata6
|
spermatogenesis associated 6 |
| chr25_-_36370292 | 0.08 |
ENSDART00000152766
|
CR354435.1
|
Histone H2B 1/2 |
| chr23_+_41800052 | 0.08 |
ENSDART00000141484
|
pdyn
|
prodynorphin |
| chr20_-_54377933 | 0.08 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr1_-_20271138 | 0.08 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr11_-_2594045 | 0.07 |
ENSDART00000114079
|
nab2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr25_-_36310561 | 0.07 |
ENSDART00000181152
|
zgc:165555
|
zgc:165555 |
| chr7_-_12596727 | 0.07 |
ENSDART00000186413
|
adamtsl3
|
ADAMTS-like 3 |
| chr15_-_14486534 | 0.07 |
ENSDART00000179368
|
numbl
|
numb homolog (Drosophila)-like |
| chr3_+_23737795 | 0.07 |
ENSDART00000182247
|
hoxb3a
|
homeobox B3a |
| chr4_-_9586713 | 0.07 |
ENSDART00000145613
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr17_+_20639180 | 0.07 |
ENSDART00000063624
|
npy4r
|
neuropeptide Y receptor Y4 |
| chr2_+_26288301 | 0.07 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr19_+_43504480 | 0.07 |
ENSDART00000159421
|
BX649384.3
|
|
| chr7_+_20017211 | 0.07 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr19_-_7225060 | 0.07 |
ENSDART00000133179
ENSDART00000151138 ENSDART00000104799 ENSDART00000128331 |
col11a2
|
collagen, type XI, alpha 2 |
| chr12_+_34273240 | 0.07 |
ENSDART00000037904
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr1_-_44940830 | 0.07 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr11_+_45461853 | 0.06 |
ENSDART00000173409
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr1_+_11659861 | 0.06 |
ENSDART00000054787
|
BX569798.1
|
|
| chr17_+_8323348 | 0.06 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr23_-_22072460 | 0.06 |
ENSDART00000132343
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr11_-_44347587 | 0.06 |
ENSDART00000172919
|
si:cabz01100188.1
|
si:cabz01100188.1 |
| chr25_+_35706493 | 0.06 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr5_-_12093618 | 0.06 |
ENSDART00000161542
|
lrrc74b
|
leucine rich repeat containing 74B |
| chr4_+_30775376 | 0.06 |
ENSDART00000158528
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr9_-_30363770 | 0.06 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr8_+_23436411 | 0.06 |
ENSDART00000140044
|
foxp3b
|
forkhead box P3b |
| chr22_-_6499055 | 0.06 |
ENSDART00000142235
ENSDART00000183588 |
si:dkey-19a16.7
|
si:dkey-19a16.7 |
| chr12_-_44016898 | 0.06 |
ENSDART00000175304
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr1_+_56972616 | 0.06 |
ENSDART00000152812
|
si:ch211-1f22.11
|
si:ch211-1f22.11 |
| chr23_-_27547931 | 0.06 |
ENSDART00000144419
|
larp4aa
|
La ribonucleoprotein domain family, member 4Aa |
| chr25_+_34889061 | 0.06 |
ENSDART00000136226
|
spire2
|
spire-type actin nucleation factor 2 |
| chr13_+_25486608 | 0.05 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr16_+_19029297 | 0.05 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr10_+_23553533 | 0.05 |
ENSDART00000159654
|
CR847844.2
|
|
| chr1_+_1599979 | 0.05 |
ENSDART00000097626
|
urp2
|
urotensin II-related peptide |
| chr1_+_54773877 | 0.05 |
ENSDART00000129831
|
nlrc6
|
NLR family CARD domain containing 6 |
| chr15_-_12319065 | 0.05 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr22_+_24770744 | 0.05 |
ENSDART00000142882
|
si:rp71-23d18.4
|
si:rp71-23d18.4 |
| chr12_+_19030391 | 0.05 |
ENSDART00000153927
|
si:ch73-139e5.2
|
si:ch73-139e5.2 |
| chr1_+_32528097 | 0.05 |
ENSDART00000128317
|
nlgn4a
|
neuroligin 4a |
| chr1_+_49435017 | 0.05 |
ENSDART00000124833
|
pdcd11
|
programmed cell death 11 |
| chr12_+_499881 | 0.05 |
ENSDART00000167527
|
mprip
|
myosin phosphatase Rho interacting protein |
| chr1_-_56363108 | 0.05 |
ENSDART00000021878
|
gcgrb
|
glucagon receptor b |
Network of associatons between targets according to the STRING database.
First level regulatory network of egr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.1 | 0.4 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.3 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.1 | 0.5 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 0.5 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.2 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 0.6 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.2 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 0.5 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.1 | 0.2 | GO:0044785 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 0.2 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.3 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.9 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.4 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.7 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.1 | GO:0060468 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.0 | 0.2 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.4 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.2 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.6 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.4 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.1 | 0.6 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.5 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.0 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |