Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
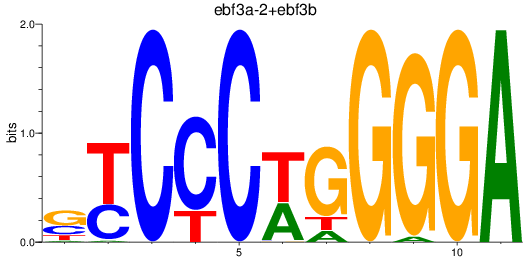
Results for ebf3a-2+ebf3b
Z-value: 0.38
Transcription factors associated with ebf3a-2+ebf3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ebf3b
|
ENSDARG00000091287 | EBF transcription factor 3b |
|
ebf3a-2
|
ENSDARG00000100244 | EBF transcription factor 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ebf3a | dr11_v1_chr12_+_42574148_42574148 | 0.30 | 2.2e-01 | Click! |
| ebf3b | dr11_v1_chr17_-_20229812_20229812 | 0.02 | 9.5e-01 | Click! |
Activity profile of ebf3a-2+ebf3b motif
Sorted Z-values of ebf3a-2+ebf3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_11756040 | 0.65 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr22_+_11775269 | 0.37 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr19_-_8880688 | 0.34 |
ENSDART00000039629
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr25_+_4541211 | 0.31 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr22_-_20761759 | 0.31 |
ENSDART00000013803
|
amh
|
anti-Mullerian hormone |
| chr13_-_4018888 | 0.30 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr16_-_32013913 | 0.23 |
ENSDART00000030282
ENSDART00000138701 |
gstk1
|
glutathione S-transferase kappa 1 |
| chr19_+_19786117 | 0.21 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr15_+_37559570 | 0.21 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr17_-_30521043 | 0.20 |
ENSDART00000087111
|
itsn2b
|
intersectin 2b |
| chr6_+_7250824 | 0.20 |
ENSDART00000177226
|
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr14_-_36397768 | 0.20 |
ENSDART00000185199
ENSDART00000052562 |
spata4
|
spermatogenesis associated 4 |
| chr9_-_36924388 | 0.19 |
ENSDART00000059756
|
ralba
|
v-ral simian leukemia viral oncogene homolog Ba (ras related) |
| chr10_-_42237304 | 0.19 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr17_+_17955063 | 0.19 |
ENSDART00000104999
|
ccdc85ca
|
coiled-coil domain containing 85C, a |
| chr22_-_18387059 | 0.19 |
ENSDART00000007769
|
tssk6
|
testis-specific serine kinase 6 |
| chr3_-_3496738 | 0.18 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr5_-_19052184 | 0.18 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr25_-_21716326 | 0.18 |
ENSDART00000152011
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr20_+_25711718 | 0.17 |
ENSDART00000033436
|
cep135
|
centrosomal protein 135 |
| chr11_-_25324534 | 0.17 |
ENSDART00000158598
|
si:ch211-232b12.5
|
si:ch211-232b12.5 |
| chr6_+_54711306 | 0.17 |
ENSDART00000074605
|
pkp1b
|
plakophilin 1b |
| chr11_-_3308569 | 0.16 |
ENSDART00000036581
|
cdk2
|
cyclin-dependent kinase 2 |
| chr1_+_15062 | 0.16 |
ENSDART00000169005
|
cep97
|
centrosomal protein 97 |
| chr13_-_31452516 | 0.16 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr12_+_34763795 | 0.16 |
ENSDART00000153097
|
slc38a10
|
solute carrier family 38, member 10 |
| chr4_+_37992753 | 0.16 |
ENSDART00000193890
|
CR759843.2
|
|
| chr20_+_18209895 | 0.15 |
ENSDART00000111063
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr6_+_45932276 | 0.14 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr2_-_37465517 | 0.14 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr10_+_31809226 | 0.14 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr2_-_10563576 | 0.14 |
ENSDART00000185818
ENSDART00000190887 |
ccdc18
|
coiled-coil domain containing 18 |
| chr13_+_17694845 | 0.13 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr3_-_55328548 | 0.12 |
ENSDART00000082944
|
dock6
|
dedicator of cytokinesis 6 |
| chr16_-_36834505 | 0.12 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr11_-_22303678 | 0.12 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr1_-_191971 | 0.12 |
ENSDART00000164091
ENSDART00000159237 |
grtp1a
|
growth hormone regulated TBC protein 1a |
| chr17_+_24222190 | 0.12 |
ENSDART00000181698
ENSDART00000189411 |
ehbp1
|
EH domain binding protein 1 |
| chr23_-_36003282 | 0.12 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr4_-_760560 | 0.11 |
ENSDART00000103601
|
agbl5
|
ATP/GTP binding protein-like 5 |
| chr11_+_3308656 | 0.11 |
ENSDART00000082458
|
sarnp
|
SAP domain containing ribonucleoprotein |
| chr7_+_17063761 | 0.11 |
ENSDART00000182880
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr2_-_55317035 | 0.11 |
ENSDART00000169382
ENSDART00000097874 |
tpm4b
|
tropomyosin 4b |
| chr24_+_32472155 | 0.11 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr7_-_33949246 | 0.10 |
ENSDART00000173513
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr20_+_38201644 | 0.10 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr8_-_21988833 | 0.10 |
ENSDART00000167708
|
nphp4
|
nephronophthisis 4 |
| chr7_+_38811800 | 0.10 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr1_-_35924495 | 0.10 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr23_-_36418059 | 0.10 |
ENSDART00000135232
|
znf740b
|
zinc finger protein 740b |
| chr19_+_10979983 | 0.10 |
ENSDART00000142975
|
si:ch1073-70f20.1
|
si:ch1073-70f20.1 |
| chr6_+_40794015 | 0.10 |
ENSDART00000144479
|
gata2b
|
GATA binding protein 2b |
| chr19_-_10043142 | 0.10 |
ENSDART00000193016
|
grin2da
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a |
| chr1_+_36911471 | 0.09 |
ENSDART00000148640
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr24_+_27023616 | 0.09 |
ENSDART00000089541
|
dip2ca
|
disco-interacting protein 2 homolog Ca |
| chr15_-_1941042 | 0.09 |
ENSDART00000112946
ENSDART00000155504 |
dock10
|
dedicator of cytokinesis 10 |
| chr16_-_34212912 | 0.08 |
ENSDART00000145017
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr24_+_25794318 | 0.08 |
ENSDART00000143395
|
sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr24_+_3972260 | 0.08 |
ENSDART00000131753
|
pfkpa
|
phosphofructokinase, platelet a |
| chr20_+_6590220 | 0.08 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr13_+_502230 | 0.08 |
ENSDART00000013007
|
degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr10_-_27199135 | 0.08 |
ENSDART00000189511
ENSDART00000180314 |
auts2a
|
autism susceptibility candidate 2a |
| chr9_-_98982 | 0.08 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr10_-_5022176 | 0.08 |
ENSDART00000148277
|
hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr16_-_30927764 | 0.07 |
ENSDART00000156804
|
slc45a4
|
solute carrier family 45, member 4 |
| chr5_-_25174420 | 0.07 |
ENSDART00000141554
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr16_-_47245563 | 0.07 |
ENSDART00000190225
|
LO017877.2
|
|
| chr6_+_102506 | 0.07 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr3_+_13559199 | 0.07 |
ENSDART00000166547
|
si:ch73-106n3.1
|
si:ch73-106n3.1 |
| chr5_-_31773208 | 0.07 |
ENSDART00000137556
ENSDART00000122066 |
fam102ab
|
family with sequence similarity 102, member Ab |
| chr19_+_9459050 | 0.07 |
ENSDART00000186419
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr9_-_33081781 | 0.07 |
ENSDART00000165748
|
zgc:172053
|
zgc:172053 |
| chr17_+_25955003 | 0.07 |
ENSDART00000156029
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr23_+_19814371 | 0.07 |
ENSDART00000182897
|
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr9_+_56443416 | 0.07 |
ENSDART00000167947
|
CABZ01079480.1
|
|
| chr20_+_46371458 | 0.07 |
ENSDART00000152912
|
adgrg11
|
adhesion G protein-coupled receptor G11 |
| chr8_-_33154677 | 0.07 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr16_-_35952789 | 0.06 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr12_-_46176115 | 0.06 |
ENSDART00000152848
|
si:ch211-226h7.8
|
si:ch211-226h7.8 |
| chr17_+_53425730 | 0.06 |
ENSDART00000127617
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr20_-_53321499 | 0.06 |
ENSDART00000179894
ENSDART00000127427 ENSDART00000084952 |
wasf1
|
WAS protein family, member 1 |
| chr17_+_26352372 | 0.06 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr17_-_22324727 | 0.06 |
ENSDART00000160341
|
CU104709.1
|
|
| chr13_+_33368140 | 0.06 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr8_+_30671060 | 0.06 |
ENSDART00000193749
|
adora2aa
|
adenosine A2a receptor a |
| chr23_+_35672542 | 0.06 |
ENSDART00000046268
|
pmelb
|
premelanosome protein b |
| chr9_+_32930622 | 0.06 |
ENSDART00000100928
|
cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr4_+_29344957 | 0.05 |
ENSDART00000185164
|
CR847895.1
|
|
| chr3_-_28665291 | 0.05 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr15_-_23936276 | 0.05 |
ENSDART00000137569
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr25_-_20523927 | 0.05 |
ENSDART00000114448
|
si:ch211-269c21.2
|
si:ch211-269c21.2 |
| chr11_+_13071645 | 0.05 |
ENSDART00000162259
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr2_+_38608290 | 0.05 |
ENSDART00000159066
|
cdh24b
|
cadherin 24, type 2b |
| chr2_-_30987538 | 0.04 |
ENSDART00000148157
|
emilin2a
|
elastin microfibril interfacer 2a |
| chr4_-_30712588 | 0.04 |
ENSDART00000142393
|
si:dkey-16p19.5
|
si:dkey-16p19.5 |
| chr17_+_45454943 | 0.04 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr25_+_31323978 | 0.04 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr25_+_34915576 | 0.04 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr25_-_13549577 | 0.04 |
ENSDART00000166772
|
ano10b
|
anoctamin 10b |
| chr7_+_39410180 | 0.04 |
ENSDART00000168641
|
CT030188.1
|
|
| chr19_+_1370504 | 0.03 |
ENSDART00000158946
|
dgat1a
|
diacylglycerol O-acyltransferase 1a |
| chr25_+_9003230 | 0.03 |
ENSDART00000180330
ENSDART00000142917 |
rag1
|
recombination activating gene 1 |
| chr3_-_55649319 | 0.03 |
ENSDART00000176127
|
axin2
|
axin 2 (conductin, axil) |
| chr13_-_40316367 | 0.03 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr4_-_38259998 | 0.03 |
ENSDART00000172389
|
znf1093
|
zinc finger protein 1093 |
| chr6_+_55277419 | 0.03 |
ENSDART00000083670
|
CABZ01041604.1
|
|
| chr4_-_28958601 | 0.03 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr2_+_37110504 | 0.03 |
ENSDART00000132794
ENSDART00000042974 |
slc1a8b
|
solute carrier family 1 (glutamate transporter), member 8b |
| chr6_-_42036685 | 0.03 |
ENSDART00000155174
|
GPR62 (1 of many)
|
si:dkeyp-111e5.4 |
| chr21_-_37194365 | 0.03 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr24_-_13364311 | 0.03 |
ENSDART00000183808
|
KCNB2
|
si:dkey-192j17.1 |
| chr10_-_5016997 | 0.03 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr23_+_16948049 | 0.02 |
ENSDART00000139449
|
si:dkey-147f3.4
|
si:dkey-147f3.4 |
| chr15_+_1372343 | 0.02 |
ENSDART00000152285
|
schip1
|
schwannomin interacting protein 1 |
| chr2_-_32457919 | 0.02 |
ENSDART00000132792
ENSDART00000041319 |
slc4a2a
|
solute carrier family 4 (anion exchanger), member 2a |
| chr8_-_25422186 | 0.02 |
ENSDART00000113492
ENSDART00000131736 |
kcnq2a
|
potassium voltage-gated channel, KQT-like subfamily, member 2a |
| chr4_-_69127091 | 0.02 |
ENSDART00000136092
|
si:ch211-209j12.3
|
si:ch211-209j12.3 |
| chr2_-_10943093 | 0.02 |
ENSDART00000148999
|
ssbp3a
|
single stranded DNA binding protein 3a |
| chr16_-_47483142 | 0.02 |
ENSDART00000147072
|
cthrc1b
|
collagen triple helix repeat containing 1b |
| chr11_-_27501027 | 0.02 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr18_+_40471826 | 0.02 |
ENSDART00000098806
|
ugt5c3
|
UDP glucuronosyltransferase 5 family, polypeptide C3 |
| chr10_+_158590 | 0.02 |
ENSDART00000081982
|
KCNJ15
|
potassium voltage-gated channel subfamily J member 15 |
| chr24_+_7800486 | 0.02 |
ENSDART00000145504
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr1_+_56873359 | 0.02 |
ENSDART00000152713
|
si:ch211-152f2.2
|
si:ch211-152f2.2 |
| chr8_-_1051438 | 0.02 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr3_-_25813426 | 0.01 |
ENSDART00000039482
|
ntn1b
|
netrin 1b |
| chr13_-_13751017 | 0.01 |
ENSDART00000180182
|
ky
|
kyphoscoliosis peptidase |
| chr23_-_5683147 | 0.01 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr14_+_22129096 | 0.01 |
ENSDART00000132514
|
ccng1
|
cyclin G1 |
| chr6_-_40697585 | 0.01 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr15_+_35718354 | 0.01 |
ENSDART00000110003
|
nyap2a
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2a |
| chr17_+_38566717 | 0.01 |
ENSDART00000145147
|
sptb
|
spectrin, beta, erythrocytic |
| chr9_-_7673856 | 0.01 |
ENSDART00000102715
|
tuba8l3
|
tubulin, alpha 8 like 3 |
| chr6_-_49673476 | 0.01 |
ENSDART00000112226
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr13_+_17702522 | 0.01 |
ENSDART00000057913
|
si:dkey-27m7.4
|
si:dkey-27m7.4 |
| chr7_-_52558495 | 0.01 |
ENSDART00000138263
ENSDART00000009938 ENSDART00000174292 ENSDART00000174218 ENSDART00000174335 |
tcf12
|
transcription factor 12 |
| chr2_-_144393 | 0.01 |
ENSDART00000156008
|
adcy1b
|
adenylate cyclase 1b |
| chr21_-_37194839 | 0.01 |
ENSDART00000175126
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr5_+_32141790 | 0.01 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chr5_-_31772559 | 0.01 |
ENSDART00000183879
|
fam102ab
|
family with sequence similarity 102, member Ab |
| chr4_+_18782840 | 0.01 |
ENSDART00000146450
|
slc26a4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr13_+_17702853 | 0.01 |
ENSDART00000145438
|
si:dkey-27m7.4
|
si:dkey-27m7.4 |
| chr16_-_36093776 | 0.01 |
ENSDART00000159134
|
OSCP1
|
organic solute carrier partner 1 |
| chr5_+_23634665 | 0.01 |
ENSDART00000125161
|
cx39.9
|
connexin 39.9 |
| chr24_-_27400017 | 0.01 |
ENSDART00000145829
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr3_+_50310684 | 0.01 |
ENSDART00000112152
|
gas7a
|
growth arrest-specific 7a |
| chr22_-_23253481 | 0.01 |
ENSDART00000054807
|
lhx9
|
LIM homeobox 9 |
| chr7_+_39410393 | 0.01 |
ENSDART00000158561
ENSDART00000185173 |
CT030188.1
|
|
| chr23_+_21459263 | 0.01 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr20_+_2039518 | 0.01 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr17_-_33407843 | 0.01 |
ENSDART00000129416
|
BX323819.1
|
|
| chr21_-_37194669 | 0.01 |
ENSDART00000192748
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr18_+_6054816 | 0.01 |
ENSDART00000113668
|
si:ch73-386h18.1
|
si:ch73-386h18.1 |
| chr24_+_1294176 | 0.01 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr21_-_24632778 | 0.01 |
ENSDART00000132533
ENSDART00000058370 |
arhgap32b
|
Rho GTPase activating protein 32b |
| chr3_+_4266289 | 0.00 |
ENSDART00000101636
|
si:dkey-73p2.1
|
si:dkey-73p2.1 |
| chr23_-_1144425 | 0.00 |
ENSDART00000098271
|
psmb11b
|
proteasome subunit beta11b |
| chr4_+_5193198 | 0.00 |
ENSDART00000067388
|
fgf23
|
fibroblast growth factor 23 |
| chr18_+_30441740 | 0.00 |
ENSDART00000189074
|
gse1
|
Gse1 coiled-coil protein |
| chr25_+_9003879 | 0.00 |
ENSDART00000186228
|
rag1
|
recombination activating gene 1 |
| chr15_+_16521785 | 0.00 |
ENSDART00000062191
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr13_-_31622195 | 0.00 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr18_+_15616167 | 0.00 |
ENSDART00000080454
|
slc17a8
|
solute carrier family 17 (vesicular glutamate transporter), member 8 |
| chr5_-_29112956 | 0.00 |
ENSDART00000132726
|
whrnb
|
whirlin b |
| chr17_+_25187226 | 0.00 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr8_+_8196087 | 0.00 |
ENSDART00000026965
|
plxnb3
|
plexin B3 |
| chr3_+_4113551 | 0.00 |
ENSDART00000192309
|
LO018551.1
|
|
| chr24_+_4977862 | 0.00 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr17_+_25187670 | 0.00 |
ENSDART00000190873
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr7_+_7630409 | 0.00 |
ENSDART00000172934
|
clcn3
|
chloride channel 3 |
| chr20_+_34933183 | 0.00 |
ENSDART00000062738
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr8_+_26293673 | 0.00 |
ENSDART00000144977
|
mgll
|
monoglyceride lipase |
Network of associatons between targets according to the STRING database.
First level regulatory network of ebf3a-2+ebf3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.1 | 0.3 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.0 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |