Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for ebf1b
Z-value: 0.78
Transcription factors associated with ebf1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ebf1b
|
ENSDARG00000069196 | EBF transcription factor 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ebf1b | dr11_v1_chr21_-_33851877_33851877 | 0.54 | 2.0e-02 | Click! |
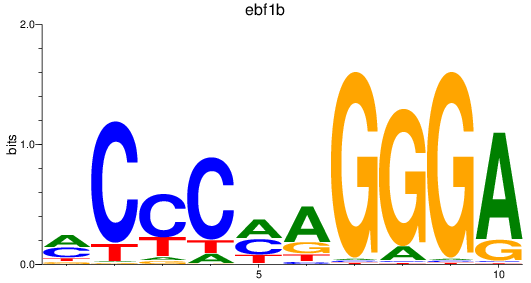
Activity profile of ebf1b motif
Sorted Z-values of ebf1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_8208464 | 3.50 |
ENSDART00000161283
|
pimr203
|
Pim proto-oncogene, serine/threonine kinase, related 203 |
| chr22_+_36914636 | 1.68 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr1_-_44701313 | 1.66 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr11_-_8271374 | 1.33 |
ENSDART00000168253
|
pimr202
|
Pim proto-oncogene, serine/threonine kinase, related 202 |
| chr11_-_8288925 | 1.25 |
ENSDART00000168524
|
pimr202
|
Pim proto-oncogene, serine/threonine kinase, related 202 |
| chr24_-_28893251 | 1.10 |
ENSDART00000042065
ENSDART00000003503 |
col11a1a
|
collagen, type XI, alpha 1a |
| chr22_+_11756040 | 1.10 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr22_-_36934040 | 1.07 |
ENSDART00000151666
|
pimr206
|
Pim proto-oncogene, serine/threonine kinase, related 206 |
| chr18_-_4957022 | 0.92 |
ENSDART00000101686
|
zbbx
|
zinc finger, B-box domain containing |
| chr1_-_9114936 | 0.88 |
ENSDART00000163624
ENSDART00000132910 |
si:ch211-14k19.8
vap
|
si:ch211-14k19.8 vascular associated protein |
| chr25_+_4960509 | 0.87 |
ENSDART00000155984
|
si:ch73-265h17.4
|
si:ch73-265h17.4 |
| chr11_+_16137302 | 0.80 |
ENSDART00000164892
|
pimr204
|
Pim proto-oncogene, serine/threonine kinase, related 204 |
| chr24_+_23730061 | 0.79 |
ENSDART00000080343
|
ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr11_-_40128722 | 0.78 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr4_+_37992753 | 0.74 |
ENSDART00000193890
|
CR759843.2
|
|
| chr23_+_45027263 | 0.73 |
ENSDART00000058364
|
hmgb2b
|
high mobility group box 2b |
| chr3_+_62317990 | 0.68 |
ENSDART00000058719
|
znf1124
|
zinc finger protein 1124 |
| chrM_+_3803 | 0.64 |
ENSDART00000093596
|
mt-nd1
|
NADH dehydrogenase 1, mitochondrial |
| chr5_-_6561376 | 0.63 |
ENSDART00000099570
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr19_-_13733870 | 0.62 |
ENSDART00000177773
|
epb41a
|
erythrocyte membrane protein band 4.1a |
| chr9_+_44431174 | 0.62 |
ENSDART00000149726
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr18_+_10820430 | 0.59 |
ENSDART00000161990
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr10_-_31563049 | 0.58 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr5_+_457729 | 0.58 |
ENSDART00000025183
|
ift74
|
intraflagellar transport 74 |
| chr4_+_11375894 | 0.57 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr8_+_31872992 | 0.57 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr22_-_18387059 | 0.55 |
ENSDART00000007769
|
tssk6
|
testis-specific serine kinase 6 |
| chr6_-_43922813 | 0.55 |
ENSDART00000123341
|
prok2
|
prokineticin 2 |
| chr3_-_3496738 | 0.55 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr2_-_43123949 | 0.55 |
ENSDART00000075347
ENSDART00000132346 |
lrrc6
|
leucine rich repeat containing 6 |
| chr6_+_11850359 | 0.53 |
ENSDART00000109552
ENSDART00000188139 ENSDART00000181499 ENSDART00000178269 |
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr12_+_47945664 | 0.52 |
ENSDART00000189824
ENSDART00000055743 ENSDART00000188936 |
CABZ01074994.1
|
|
| chr10_+_33982010 | 0.51 |
ENSDART00000180431
|
fryb
|
furry homolog b (Drosophila) |
| chr7_+_61876641 | 0.49 |
ENSDART00000165409
|
CR382383.2
|
|
| chr8_+_12930216 | 0.49 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr19_+_40335271 | 0.49 |
ENSDART00000135648
|
fam133b
|
family with sequence similarity 133, member B |
| chr11_-_8782871 | 0.48 |
ENSDART00000158546
|
si:ch211-51h4.2
|
si:ch211-51h4.2 |
| chr11_+_8660158 | 0.47 |
ENSDART00000169141
|
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr23_+_30898013 | 0.47 |
ENSDART00000146859
|
cables2a
|
Cdk5 and Abl enzyme substrate 2a |
| chr8_-_34762163 | 0.45 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr10_-_8041948 | 0.44 |
ENSDART00000059017
|
nfkbil1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr10_-_27196093 | 0.43 |
ENSDART00000185282
|
auts2a
|
autism susceptibility candidate 2a |
| chr9_-_22831836 | 0.43 |
ENSDART00000142585
|
neb
|
nebulin |
| chr1_-_35924495 | 0.43 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr8_+_30671060 | 0.40 |
ENSDART00000193749
|
adora2aa
|
adenosine A2a receptor a |
| chr10_-_9115383 | 0.40 |
ENSDART00000139324
|
si:dkeyp-41f9.3
|
si:dkeyp-41f9.3 |
| chr7_+_25920792 | 0.39 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr20_+_46371458 | 0.37 |
ENSDART00000152912
|
adgrg11
|
adhesion G protein-coupled receptor G11 |
| chr9_-_45149695 | 0.36 |
ENSDART00000162033
|
CU467837.1
|
|
| chr18_+_45504362 | 0.35 |
ENSDART00000140089
|
cngb1a
|
cyclic nucleotide gated channel beta 1a |
| chr3_-_46403778 | 0.34 |
ENSDART00000074422
|
cdip1
|
cell death-inducing p53 target 1 |
| chr4_+_11695979 | 0.34 |
ENSDART00000137736
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr18_-_13360106 | 0.33 |
ENSDART00000091512
|
cmip
|
c-Maf inducing protein |
| chr16_-_30927764 | 0.32 |
ENSDART00000156804
|
slc45a4
|
solute carrier family 45, member 4 |
| chr3_+_13559199 | 0.32 |
ENSDART00000166547
|
si:ch73-106n3.1
|
si:ch73-106n3.1 |
| chr24_+_3050020 | 0.31 |
ENSDART00000004241
|
inhbaa
|
inhibin, beta Aa |
| chr22_+_567733 | 0.30 |
ENSDART00000171036
|
usp49
|
ubiquitin specific peptidase 49 |
| chr17_-_24364527 | 0.30 |
ENSDART00000155192
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr3_-_38783951 | 0.30 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr23_+_40460333 | 0.29 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr7_-_52558495 | 0.28 |
ENSDART00000138263
ENSDART00000009938 ENSDART00000174292 ENSDART00000174218 ENSDART00000174335 |
tcf12
|
transcription factor 12 |
| chr3_-_30158395 | 0.28 |
ENSDART00000103502
|
si:ch211-152f23.5
|
si:ch211-152f23.5 |
| chr17_+_24222190 | 0.28 |
ENSDART00000181698
ENSDART00000189411 |
ehbp1
|
EH domain binding protein 1 |
| chr16_-_14534724 | 0.26 |
ENSDART00000148126
|
si:dkey-237j11.3
|
si:dkey-237j11.3 |
| chr23_+_2760573 | 0.25 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr20_+_25711718 | 0.24 |
ENSDART00000033436
|
cep135
|
centrosomal protein 135 |
| chr11_-_22303678 | 0.24 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr4_+_76745015 | 0.24 |
ENSDART00000155883
|
ms4a17a.10
|
membrane-spanning 4-domains, subfamily A, member 17A.10 |
| chr4_+_6840223 | 0.23 |
ENSDART00000161329
|
dock4b
|
dedicator of cytokinesis 4b |
| chr15_+_11427620 | 0.23 |
ENSDART00000168688
|
ndufc2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 |
| chr15_-_30505607 | 0.23 |
ENSDART00000155212
|
msi2b
|
musashi RNA-binding protein 2b |
| chr24_-_25184553 | 0.22 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr16_-_21785261 | 0.22 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr13_-_32906265 | 0.21 |
ENSDART00000113823
ENSDART00000171114 ENSDART00000180035 ENSDART00000137570 |
si:dkey-18j18.3
|
si:dkey-18j18.3 |
| chr3_-_3209432 | 0.21 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr7_-_52842007 | 0.20 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr2_+_56891858 | 0.20 |
ENSDART00000159912
|
zgc:85843
|
zgc:85843 |
| chr14_-_21959712 | 0.20 |
ENSDART00000021417
|
p2rx3a
|
purinergic receptor P2X, ligand-gated ion channel, 3a |
| chr16_-_33076764 | 0.19 |
ENSDART00000192917
|
dopey1
|
dopey family member 1 |
| chr7_+_58686860 | 0.19 |
ENSDART00000052332
|
penkb
|
proenkephalin b |
| chr9_-_1200187 | 0.19 |
ENSDART00000158760
|
ino80da
|
INO80 complex subunit Da |
| chr5_+_32141790 | 0.19 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chr1_-_16688965 | 0.19 |
ENSDART00000079001
ENSDART00000178599 |
frg1
|
FSHD region gene 1 |
| chr1_+_41666611 | 0.18 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr17_+_26352372 | 0.18 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr20_-_46467280 | 0.17 |
ENSDART00000060702
|
rmdn3
|
regulator of microtubule dynamics 3 |
| chr23_-_6920107 | 0.17 |
ENSDART00000149230
ENSDART00000182946 |
si:ch211-117c9.2
|
si:ch211-117c9.2 |
| chr19_-_26863626 | 0.17 |
ENSDART00000145568
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr11_+_25638172 | 0.17 |
ENSDART00000114226
ENSDART00000143677 |
grm6b
|
glutamate receptor, metabotropic 6b |
| chr18_-_19095141 | 0.17 |
ENSDART00000090962
ENSDART00000090922 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr18_+_33580391 | 0.16 |
ENSDART00000189300
|
si:dkey-47k20.4
|
si:dkey-47k20.4 |
| chr19_-_35400819 | 0.16 |
ENSDART00000148080
|
rnf19b
|
ring finger protein 19B |
| chr16_+_12281032 | 0.15 |
ENSDART00000138638
|
si:dkey-26c10.5
|
si:dkey-26c10.5 |
| chr7_-_41693004 | 0.15 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr25_+_21324588 | 0.15 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr9_+_44430974 | 0.15 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr22_-_28373698 | 0.14 |
ENSDART00000157592
|
si:ch211-213c4.5
|
si:ch211-213c4.5 |
| chr16_-_47245563 | 0.14 |
ENSDART00000190225
|
LO017877.2
|
|
| chr25_+_31323978 | 0.14 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr22_+_39058269 | 0.14 |
ENSDART00000113362
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr13_+_17702853 | 0.13 |
ENSDART00000145438
|
si:dkey-27m7.4
|
si:dkey-27m7.4 |
| chr18_+_20871142 | 0.13 |
ENSDART00000188274
|
pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr14_+_7898372 | 0.13 |
ENSDART00000159593
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr18_-_15551360 | 0.13 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr2_+_22495274 | 0.13 |
ENSDART00000167915
|
lrrc8da
|
leucine rich repeat containing 8 VRAC subunit Da |
| chr4_-_10599062 | 0.12 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr2_+_38608290 | 0.12 |
ENSDART00000159066
|
cdh24b
|
cadherin 24, type 2b |
| chr4_-_5019113 | 0.12 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr14_-_41369629 | 0.11 |
ENSDART00000173040
|
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr3_+_3545825 | 0.11 |
ENSDART00000109060
|
CR589947.1
|
|
| chr5_+_36932718 | 0.10 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr15_-_6976851 | 0.10 |
ENSDART00000158474
ENSDART00000168943 ENSDART00000169944 |
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr13_+_17702522 | 0.10 |
ENSDART00000057913
|
si:dkey-27m7.4
|
si:dkey-27m7.4 |
| chr21_-_14775724 | 0.10 |
ENSDART00000043239
|
glulc
|
glutamate-ammonia ligase (glutamine synthase) c |
| chr25_+_35212919 | 0.09 |
ENSDART00000180127
|
ano5a
|
anoctamin 5a |
| chr4_+_20318127 | 0.09 |
ENSDART00000028856
ENSDART00000132909 |
cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr9_+_36356740 | 0.09 |
ENSDART00000139033
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr5_+_9360394 | 0.08 |
ENSDART00000124642
|
FP236810.2
|
|
| chr24_-_39826865 | 0.08 |
ENSDART00000089232
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr3_+_36160979 | 0.08 |
ENSDART00000038525
|
si:ch211-234h8.7
|
si:ch211-234h8.7 |
| chr20_+_48782068 | 0.08 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr4_+_74255904 | 0.07 |
ENSDART00000174385
|
ptprr
|
protein tyrosine phosphatase, receptor type, r |
| chr1_-_18772241 | 0.07 |
ENSDART00000147645
ENSDART00000133194 ENSDART00000147525 ENSDART00000191493 |
tbc1d1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr24_+_27023616 | 0.06 |
ENSDART00000089541
|
dip2ca
|
disco-interacting protein 2 homolog Ca |
| chr1_+_41690402 | 0.06 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr16_-_25535430 | 0.06 |
ENSDART00000077420
|
dtnbp1a
|
dystrobrevin binding protein 1a |
| chr21_-_19018455 | 0.06 |
ENSDART00000080256
|
nefma
|
neurofilament, medium polypeptide a |
| chr22_+_28446365 | 0.06 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr25_+_19666621 | 0.06 |
ENSDART00000112746
|
zgc:171459
|
zgc:171459 |
| chr15_-_23420079 | 0.06 |
ENSDART00000104525
ENSDART00000148840 |
kmt2a
|
lysine (K)-specific methyltransferase 2A |
| chr21_-_7687544 | 0.05 |
ENSDART00000134519
|
pde8b
|
phosphodiesterase 8B |
| chr4_-_72468168 | 0.05 |
ENSDART00000182995
ENSDART00000174067 |
CR788316.1
|
|
| chr3_-_42086577 | 0.05 |
ENSDART00000083111
ENSDART00000187312 |
ttyh3a
|
tweety family member 3a |
| chr23_-_19230627 | 0.05 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr8_+_8196087 | 0.05 |
ENSDART00000026965
|
plxnb3
|
plexin B3 |
| chr21_-_2273244 | 0.05 |
ENSDART00000171417
|
si:ch73-299h12.3
|
si:ch73-299h12.3 |
| chr9_-_34296406 | 0.05 |
ENSDART00000125451
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr21_+_26389391 | 0.04 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr24_-_25691020 | 0.04 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr23_+_19790962 | 0.04 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr11_-_33857911 | 0.04 |
ENSDART00000165370
|
nxph2b
|
neurexophilin 2b |
| chr17_+_43629008 | 0.03 |
ENSDART00000184185
ENSDART00000181681 |
znf365
|
zinc finger protein 365 |
| chr16_-_48914757 | 0.03 |
ENSDART00000166740
|
FQ976914.1
|
|
| chr21_+_16144014 | 0.03 |
ENSDART00000163267
|
qrfpr4
|
pyroglutamylated RFamide peptide receptor 4 |
| chr4_+_43408004 | 0.03 |
ENSDART00000150476
|
si:dkeyp-53e4.2
|
si:dkeyp-53e4.2 |
| chr7_+_25221757 | 0.03 |
ENSDART00000173551
|
exoc6b
|
exocyst complex component 6B |
| chr10_+_19554604 | 0.03 |
ENSDART00000063806
|
atp6v1b2
|
ATPase H+ transporting V1 subunit B2 |
| chr22_-_7809904 | 0.03 |
ENSDART00000182269
ENSDART00000097201 ENSDART00000138522 |
si:ch73-44m9.3
si:ch73-44m9.1
|
si:ch73-44m9.3 si:ch73-44m9.1 |
| chr23_-_31506854 | 0.02 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr22_+_38778854 | 0.02 |
ENSDART00000182926
ENSDART00000125466 |
alpi.2
|
alkaline phosphatase, intestinal, tandem duplicate 2 |
| chr7_-_32599669 | 0.02 |
ENSDART00000173752
|
kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr21_+_12056934 | 0.02 |
ENSDART00000125380
|
zgc:162344
|
zgc:162344 |
| chr3_-_51142942 | 0.01 |
ENSDART00000160789
|
si:ch211-148f13.1
|
si:ch211-148f13.1 |
| chr5_-_66296635 | 0.01 |
ENSDART00000125993
|
prlrb
|
prolactin receptor b |
| chr14_+_25817246 | 0.01 |
ENSDART00000136733
|
glra1
|
glycine receptor, alpha 1 |
| chr23_+_2825940 | 0.01 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr1_-_16394814 | 0.01 |
ENSDART00000013024
|
fgf20a
|
fibroblast growth factor 20a |
| chr25_-_13105310 | 0.01 |
ENSDART00000172269
|
smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr7_-_38806186 | 0.01 |
ENSDART00000173669
|
atg13
|
ATG13 autophagy related 13 homolog (S. cerevisiae) |
| chr15_-_3466015 | 0.01 |
ENSDART00000169559
|
cog6
|
component of oligomeric golgi complex 6 |
| chr10_+_21650828 | 0.01 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr2_-_32457919 | 0.01 |
ENSDART00000132792
ENSDART00000041319 |
slc4a2a
|
solute carrier family 4 (anion exchanger), member 2a |
| chr24_-_12749116 | 0.01 |
ENSDART00000093583
|
cox19
|
COX19 cytochrome c oxidase assembly factor |
| chr11_+_3575963 | 0.00 |
ENSDART00000077305
|
si:dkey-33m11.8
|
si:dkey-33m11.8 |
| chr20_+_34933183 | 0.00 |
ENSDART00000062738
|
snap25a
|
synaptosomal-associated protein, 25a |
Network of associatons between targets according to the STRING database.
First level regulatory network of ebf1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.4 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 0.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) pronephric proximal tubule development(GO:0035776) |
| 0.1 | 0.4 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.6 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.6 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.9 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.5 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 8.6 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.1 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.8 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.7 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.1 | 1.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.6 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.4 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 1.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.3 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.0 | GO:0030882 | lipid antigen binding(GO:0030882) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |