Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for drgx_arxb
Z-value: 0.33
Transcription factors associated with drgx_arxb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
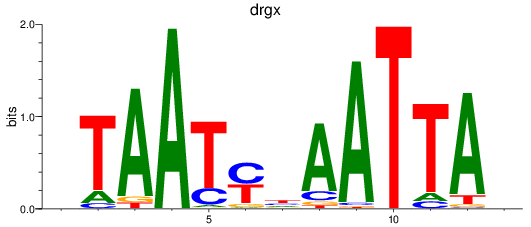
drgx
|
ENSDARG00000069329 | dorsal root ganglia homeobox |
|
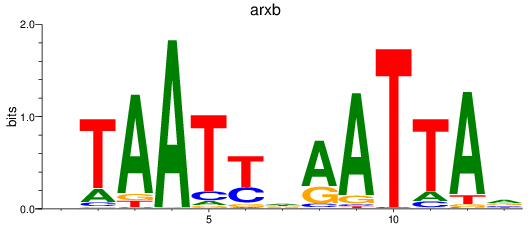
arxb
|
ENSDARG00000101523 | aristaless related homeobox b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| drgx | dr11_v1_chr13_+_30912117_30912117 | -0.90 | 5.0e-07 | Click! |
| arxb | dr11_v1_chr21_-_40676224_40676224 | -0.87 | 2.1e-06 | Click! |
Activity profile of drgx_arxb motif
Sorted Z-values of drgx_arxb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_57924611 | 1.49 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr3_-_32337653 | 1.34 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr15_-_25099679 | 1.11 |
ENSDART00000154628
|
rflnb
|
refilin B |
| chr14_-_33481428 | 1.01 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr17_-_30635298 | 0.88 |
ENSDART00000155478
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr12_-_35386910 | 0.87 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr1_-_18811517 | 0.85 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr15_-_25099233 | 0.84 |
ENSDART00000192617
|
rflnb
|
refilin B |
| chr2_-_38284648 | 0.75 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr19_-_25113660 | 0.72 |
ENSDART00000035538
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr20_-_20931197 | 0.68 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr23_-_36441693 | 0.68 |
ENSDART00000024354
|
csad
|
cysteine sulfinic acid decarboxylase |
| chr12_+_27232173 | 0.62 |
ENSDART00000193714
|
tmem106a
|
transmembrane protein 106A |
| chr11_-_3535537 | 0.59 |
ENSDART00000165329
ENSDART00000009788 |
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr7_-_37895771 | 0.58 |
ENSDART00000084282
|
papd5
|
PAP associated domain containing 5 |
| chr12_+_27231607 | 0.58 |
ENSDART00000066270
|
tmem106a
|
transmembrane protein 106A |
| chr18_-_13121983 | 0.56 |
ENSDART00000092648
|
rxylt1
|
ribitol xylosyltransferase 1 |
| chr11_+_25693395 | 0.55 |
ENSDART00000110224
|
mon1bb
|
MON1 secretory trafficking family member Bb |
| chr2_-_47957673 | 0.55 |
ENSDART00000056305
|
fzd8b
|
frizzled class receptor 8b |
| chr24_+_21346796 | 0.54 |
ENSDART00000126519
|
shisa2b
|
shisa family member 2b |
| chr20_-_20930926 | 0.54 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr12_+_23850661 | 0.49 |
ENSDART00000152921
|
svila
|
supervillin a |
| chr15_+_22435460 | 0.49 |
ENSDART00000031976
|
tmem136a
|
transmembrane protein 136a |
| chr18_+_1703984 | 0.47 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr10_+_40284003 | 0.47 |
ENSDART00000062795
ENSDART00000193825 ENSDART00000113582 |
git2b
|
G protein-coupled receptor kinase interacting ArfGAP 2b |
| chr15_+_25528290 | 0.46 |
ENSDART00000123143
|
npat
|
nuclear protein, ataxia-telangiectasia locus |
| chr15_-_17813680 | 0.46 |
ENSDART00000158556
|
CT573342.2
|
|
| chr3_-_26787430 | 0.46 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr1_-_40102836 | 0.44 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr17_-_43863700 | 0.43 |
ENSDART00000157530
|
ahsa1b
|
AHA1, activator of heat shock protein ATPase homolog 1b |
| chr11_+_31380495 | 0.41 |
ENSDART00000185073
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr14_-_34605607 | 0.41 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr17_-_31483469 | 0.39 |
ENSDART00000062907
ENSDART00000061547 |
ltk
|
leukocyte receptor tyrosine kinase |
| chr22_-_22337382 | 0.39 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr8_-_18203274 | 0.39 |
ENSDART00000134078
ENSDART00000180235 ENSDART00000080006 ENSDART00000125418 ENSDART00000142114 |
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr16_-_39267185 | 0.36 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr17_+_46387086 | 0.33 |
ENSDART00000157079
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr21_-_27881752 | 0.33 |
ENSDART00000132583
|
nrxn2a
|
neurexin 2a |
| chr25_-_13381854 | 0.32 |
ENSDART00000164621
ENSDART00000169129 |
ndrg4
|
NDRG family member 4 |
| chr17_+_28102487 | 0.29 |
ENSDART00000131638
ENSDART00000076265 |
zgc:91908
|
zgc:91908 |
| chr6_-_10708960 | 0.29 |
ENSDART00000157704
|
si:dkey-34m19.3
|
si:dkey-34m19.3 |
| chr14_-_34605804 | 0.29 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr21_+_42717424 | 0.28 |
ENSDART00000166936
ENSDART00000172135 |
sh3pxd2b
|
SH3 and PX domains 2B |
| chr7_+_71764883 | 0.27 |
ENSDART00000166865
|
myl12.1
|
myosin, light chain 12, genome duplicate 1 |
| chr15_+_12429206 | 0.27 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr7_+_29167744 | 0.27 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr9_+_29643036 | 0.26 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr5_-_32489796 | 0.26 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr5_-_69180587 | 0.24 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr25_+_31276842 | 0.24 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr2_+_37227011 | 0.24 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr12_-_47601845 | 0.23 |
ENSDART00000169548
ENSDART00000182889 |
rgs7b
|
regulator of G protein signaling 7b |
| chr18_+_3243292 | 0.23 |
ENSDART00000166580
|
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr8_+_20393250 | 0.23 |
ENSDART00000015038
|
zap70
|
zeta chain of T cell receptor associated protein kinase 70 |
| chr24_-_38079261 | 0.22 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr13_+_18507592 | 0.22 |
ENSDART00000142622
|
si:ch211-198a12.6
|
si:ch211-198a12.6 |
| chr16_+_46410520 | 0.21 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr5_-_69180227 | 0.21 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr13_+_39315881 | 0.20 |
ENSDART00000135999
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr8_-_43834442 | 0.20 |
ENSDART00000191927
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr13_-_49444636 | 0.20 |
ENSDART00000136991
|
irf2bp2a
|
interferon regulatory factor 2 binding protein 2a |
| chr6_+_8315050 | 0.20 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr13_-_25819825 | 0.20 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr25_+_33002963 | 0.20 |
ENSDART00000187366
|
CABZ01046980.1
|
|
| chr4_-_5775507 | 0.19 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr8_+_23861461 | 0.19 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr17_-_27382826 | 0.19 |
ENSDART00000186657
ENSDART00000155986 ENSDART00000191060 ENSDART00000077608 |
si:ch1073-358c10.1
|
si:ch1073-358c10.1 |
| chr15_-_5720583 | 0.18 |
ENSDART00000158034
ENSDART00000190332 ENSDART00000109599 |
urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr14_-_32631013 | 0.18 |
ENSDART00000176815
|
atp11c
|
ATPase phospholipid transporting 11C |
| chr2_-_26596794 | 0.17 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr9_+_36314867 | 0.17 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr20_+_39250673 | 0.17 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr7_+_47243564 | 0.17 |
ENSDART00000098942
ENSDART00000162237 |
znf507
|
zinc finger protein 507 |
| chr13_-_29421331 | 0.15 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr5_+_6672870 | 0.14 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr25_-_19648154 | 0.14 |
ENSDART00000148570
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr6_+_16468776 | 0.13 |
ENSDART00000109151
ENSDART00000114667 |
zgc:161969
|
zgc:161969 |
| chr9_-_21825913 | 0.13 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr17_+_21486047 | 0.13 |
ENSDART00000104608
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr18_-_17145111 | 0.13 |
ENSDART00000090446
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr6_+_39942417 | 0.12 |
ENSDART00000183256
|
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr3_-_25268751 | 0.12 |
ENSDART00000139423
|
mgat3a
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase a |
| chr17_-_1705013 | 0.12 |
ENSDART00000182864
|
CABZ01086293.1
|
|
| chr22_+_39007533 | 0.12 |
ENSDART00000185958
ENSDART00000129848 |
fam208aa
|
family with sequence similarity 208, member Aa |
| chr3_+_3610140 | 0.12 |
ENSDART00000104977
|
pick1
|
protein interacting with prkca 1 |
| chr11_+_31864921 | 0.11 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr8_+_10001805 | 0.11 |
ENSDART00000132894
|
si:dkey-8e10.2
|
si:dkey-8e10.2 |
| chr9_-_51436377 | 0.11 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr20_-_28931901 | 0.11 |
ENSDART00000153082
ENSDART00000046042 |
susd6
|
sushi domain containing 6 |
| chr13_-_31017960 | 0.11 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr8_+_50953776 | 0.11 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr8_-_43847138 | 0.11 |
ENSDART00000148358
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr13_+_15581270 | 0.10 |
ENSDART00000189880
ENSDART00000190067 ENSDART00000041293 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr21_-_18262287 | 0.10 |
ENSDART00000176716
|
vav2
|
vav 2 guanine nucleotide exchange factor |
| chr5_+_61361815 | 0.10 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr8_-_38406831 | 0.09 |
ENSDART00000112991
ENSDART00000191445 |
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr11_+_36409457 | 0.09 |
ENSDART00000077641
|
cyb561d1
|
cytochrome b561 family, member D1 |
| chr23_+_25135858 | 0.09 |
ENSDART00000103986
|
fam3a
|
family with sequence similarity 3, member A |
| chr3_-_31069776 | 0.08 |
ENSDART00000167462
|
elob
|
elongin B |
| chr6_+_40832635 | 0.08 |
ENSDART00000011931
|
ruvbl1
|
RuvB-like AAA ATPase 1 |
| chr6_+_58698475 | 0.08 |
ENSDART00000056138
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr23_-_18130264 | 0.07 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr3_+_3852614 | 0.07 |
ENSDART00000184334
ENSDART00000162313 |
si:ch73-338o16.4
|
si:ch73-338o16.4 |
| chr2_+_50967947 | 0.07 |
ENSDART00000162288
|
si:ch211-249o11.5
|
si:ch211-249o11.5 |
| chr11_-_16115804 | 0.07 |
ENSDART00000143436
ENSDART00000157928 |
rpf1
|
ribosome production factor 1 homolog |
| chr1_-_999556 | 0.07 |
ENSDART00000170884
ENSDART00000172235 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr9_-_53537989 | 0.06 |
ENSDART00000114022
|
slitrk5b
|
SLIT and NTRK-like family, member 5b |
| chr13_-_50002852 | 0.06 |
ENSDART00000099439
|
lyst
|
lysosomal trafficking regulator |
| chr22_+_17433968 | 0.06 |
ENSDART00000139523
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr1_-_23110740 | 0.06 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr17_-_14726824 | 0.06 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr13_-_9091354 | 0.06 |
ENSDART00000140319
ENSDART00000142697 |
HTRA2 (1 of many)
|
si:dkey-112g5.13 |
| chr5_-_18897482 | 0.05 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr14_+_22152060 | 0.05 |
ENSDART00000082697
ENSDART00000054982 |
gabra6a
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6a |
| chr21_+_4313039 | 0.05 |
ENSDART00000141146
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr13_-_9045879 | 0.05 |
ENSDART00000155463
ENSDART00000140041 ENSDART00000137454 |
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr17_-_12966907 | 0.05 |
ENSDART00000022874
|
psma6a
|
proteasome subunit alpha 6a |
| chr8_+_35212233 | 0.05 |
ENSDART00000111021
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr20_-_43775495 | 0.05 |
ENSDART00000100610
ENSDART00000149001 ENSDART00000148809 ENSDART00000100608 |
matn3a
|
matrilin 3a |
| chr10_-_40612773 | 0.04 |
ENSDART00000136685
ENSDART00000161373 |
taar16f
|
trace amine associated receptor 16f |
| chr10_-_40557210 | 0.04 |
ENSDART00000135297
|
taar18b
|
trace amine associated receptor 18b |
| chr12_-_34713888 | 0.04 |
ENSDART00000153272
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr13_+_411607 | 0.04 |
ENSDART00000186483
|
CU570800.3
|
|
| chr18_+_33468099 | 0.03 |
ENSDART00000131769
|
v2rh9
|
vomeronasal 2 receptor, h9 |
| chr23_+_30967686 | 0.03 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr25_+_5755314 | 0.03 |
ENSDART00000172088
ENSDART00000164208 |
appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr23_-_16484383 | 0.03 |
ENSDART00000187839
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr12_+_42436920 | 0.03 |
ENSDART00000177303
|
ebf3a
|
early B cell factor 3a |
| chr20_+_31287356 | 0.03 |
ENSDART00000007688
|
slc22a16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr2_-_20890429 | 0.03 |
ENSDART00000149989
|
pdca
|
phosducin a |
| chr21_-_10446405 | 0.03 |
ENSDART00000167948
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr14_-_41388178 | 0.03 |
ENSDART00000124532
ENSDART00000125016 ENSDART00000169247 |
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr20_+_41664350 | 0.03 |
ENSDART00000186247
|
fam184a
|
family with sequence similarity 184, member A |
| chr21_-_17482465 | 0.03 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr18_+_33171765 | 0.03 |
ENSDART00000064153
|
si:ch73-125k17.2
|
si:ch73-125k17.2 |
| chr10_-_1725699 | 0.02 |
ENSDART00000183555
|
gal3st1b
|
galactose-3-O-sulfotransferase 1b |
| chr15_+_31332552 | 0.02 |
ENSDART00000134933
ENSDART00000173915 |
or119-2
|
odorant receptor, family F, subfamily 119, member 2 |
| chr24_-_38384432 | 0.02 |
ENSDART00000140739
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr5_+_11407504 | 0.02 |
ENSDART00000186723
ENSDART00000018047 |
zgc:112294
|
zgc:112294 |
| chr14_-_17068511 | 0.02 |
ENSDART00000163766
|
phox2bb
|
paired-like homeobox 2bb |
| chr20_+_22130284 | 0.02 |
ENSDART00000025575
|
clocka
|
clock circadian regulator a |
| chr11_+_25539698 | 0.02 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr5_-_7199998 | 0.02 |
ENSDART00000167316
|
CABZ01087502.1
|
|
| chr5_-_58840971 | 0.02 |
ENSDART00000050932
|
tmem136b
|
transmembrane protein 136b |
| chr23_-_29553430 | 0.02 |
ENSDART00000157773
ENSDART00000126384 |
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr21_+_22237964 | 0.02 |
ENSDART00000003181
|
myo7ab
|
myosin VIIAb |
| chr17_+_18031304 | 0.01 |
ENSDART00000127259
|
setd3
|
SET domain containing 3 |
| chr12_+_10631266 | 0.01 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr10_-_40553066 | 0.01 |
ENSDART00000148118
ENSDART00000125958 |
taar18c
|
trace amine associated receptor 18c |
| chr14_-_17068712 | 0.01 |
ENSDART00000170277
|
phox2bb
|
paired-like homeobox 2bb |
| chr22_-_27113332 | 0.01 |
ENSDART00000146951
ENSDART00000178855 |
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr3_-_12970418 | 0.01 |
ENSDART00000158747
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr6_+_52938534 | 0.01 |
ENSDART00000174150
|
BX649282.2
|
|
| chr22_-_15693085 | 0.01 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr21_+_21357309 | 0.01 |
ENSDART00000079634
ENSDART00000146102 |
kcnk12l
|
potassium channel, subfamily K, member 12 like |
| chr2_+_20605186 | 0.01 |
ENSDART00000128505
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr2_+_49417900 | 0.01 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr10_-_33621739 | 0.01 |
ENSDART00000142655
ENSDART00000128049 |
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr15_+_45640906 | 0.01 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr5_+_40224938 | 0.01 |
ENSDART00000142897
|
si:dkey-193c22.2
|
si:dkey-193c22.2 |
| chr3_+_32594357 | 0.01 |
ENSDART00000151639
|
unc119.2
|
unc-119 lipid binding chaperone B homolog 2 |
| chr22_+_1821718 | 0.01 |
ENSDART00000132220
|
znf1002
|
zinc finger protein 1002 |
| chr16_-_27640995 | 0.01 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr21_-_44104600 | 0.01 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr24_-_7697274 | 0.00 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr10_+_23537576 | 0.00 |
ENSDART00000130515
|
CR847844.1
|
|
| chr6_+_23887314 | 0.00 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr12_+_42436328 | 0.00 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr10_+_4807913 | 0.00 |
ENSDART00000186099
|
palm2
|
paralemmin 2 |
| chr17_-_45254585 | 0.00 |
ENSDART00000185507
ENSDART00000172080 |
ttbk2a
|
tau tubulin kinase 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of drgx_arxb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 0.3 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 1.0 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.4 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.4 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.2 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 1.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.4 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.3 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.7 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.6 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.8 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 1.5 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.1 | GO:0060420 | box C/D snoRNP assembly(GO:0000492) regulation of heart growth(GO:0060420) |
| 0.0 | 0.1 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.4 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 1.3 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 2.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 1.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.0 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |