Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
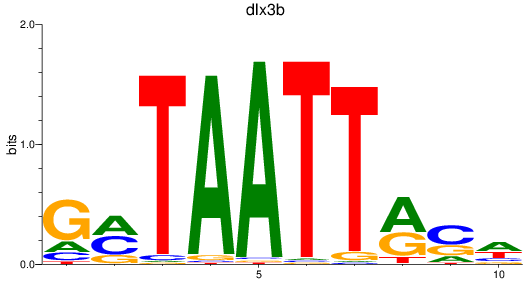
Results for dlx3b
Z-value: 0.41
Transcription factors associated with dlx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dlx3b
|
ENSDARG00000014626 | distal-less homeobox 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dlx3b | dr11_v1_chr12_+_5708400_5708400 | 0.66 | 3.0e-03 | Click! |
Activity profile of dlx3b motif
Sorted Z-values of dlx3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25777425 | 1.82 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr24_-_17029374 | 0.90 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr11_-_44801968 | 0.85 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr4_+_13586689 | 0.79 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr5_+_26213874 | 0.76 |
ENSDART00000193816
ENSDART00000098514 |
oclnb
|
occludin b |
| chr16_+_39159752 | 0.76 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr24_-_17023392 | 0.75 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr3_-_26183699 | 0.72 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr15_-_33172246 | 0.72 |
ENSDART00000158666
|
nbeab
|
neurobeachin b |
| chr12_-_35386910 | 0.66 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr11_-_39118882 | 0.59 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr10_-_21362071 | 0.54 |
ENSDART00000125167
|
avd
|
avidin |
| chr19_+_20793237 | 0.52 |
ENSDART00000014774
|
txnl4a
|
thioredoxin-like 4A |
| chr17_+_16046314 | 0.52 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr19_+_20793388 | 0.50 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr10_-_21362320 | 0.50 |
ENSDART00000189789
|
avd
|
avidin |
| chr6_-_40713183 | 0.48 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr19_+_46158078 | 0.47 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr17_-_15546862 | 0.43 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr4_+_13586455 | 0.43 |
ENSDART00000187230
|
tnpo3
|
transportin 3 |
| chr12_-_18577983 | 0.41 |
ENSDART00000193262
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr17_+_16046132 | 0.41 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr2_-_15324837 | 0.40 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr8_-_15129573 | 0.40 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr8_-_23780334 | 0.40 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr2_+_2168547 | 0.38 |
ENSDART00000029347
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr5_+_59234421 | 0.37 |
ENSDART00000045518
|
prkrip1
|
PRKR interacting protein 1 |
| chr4_+_9177997 | 0.36 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr11_-_40147032 | 0.35 |
ENSDART00000052918
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr19_-_19871211 | 0.35 |
ENSDART00000170980
|
evx1
|
even-skipped homeobox 1 |
| chr8_+_28452738 | 0.35 |
ENSDART00000062706
|
tmem189
|
transmembrane protein 189 |
| chr13_+_38814521 | 0.32 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr22_-_14262115 | 0.31 |
ENSDART00000168264
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr11_+_31864921 | 0.30 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr11_+_18873619 | 0.29 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr8_-_39822917 | 0.29 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr17_+_49500820 | 0.29 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr25_+_28555584 | 0.29 |
ENSDART00000157046
|
SLC15A5
|
si:ch211-190o6.3 |
| chr16_-_13623659 | 0.29 |
ENSDART00000168978
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr23_-_18130264 | 0.28 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chrM_+_9052 | 0.28 |
ENSDART00000093612
|
mt-atp6
|
ATP synthase 6, mitochondrial |
| chr24_-_25004553 | 0.27 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr19_-_8768564 | 0.27 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr6_-_50203682 | 0.26 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr3_+_32832538 | 0.26 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr15_-_18115540 | 0.26 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr22_-_3564563 | 0.26 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr2_-_28671139 | 0.26 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr19_-_20403845 | 0.25 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr15_-_9272328 | 0.25 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr8_-_23776399 | 0.25 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr20_+_28803977 | 0.25 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr3_-_21280373 | 0.24 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr16_-_21620947 | 0.24 |
ENSDART00000115011
ENSDART00000183125 ENSDART00000188856 ENSDART00000189460 |
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr7_-_69298271 | 0.23 |
ENSDART00000122935
|
phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr8_-_5847533 | 0.23 |
ENSDART00000192489
|
CABZ01102147.1
|
|
| chr13_+_38817871 | 0.23 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr1_+_37752171 | 0.22 |
ENSDART00000183247
ENSDART00000189756 ENSDART00000139448 |
GALNTL6
|
si:ch211-15e22.3 |
| chr14_-_32631013 | 0.22 |
ENSDART00000176815
|
atp11c
|
ATPase phospholipid transporting 11C |
| chr18_+_41232719 | 0.22 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr5_+_22067570 | 0.22 |
ENSDART00000045574
|
shisa2a
|
shisa family member 2a |
| chr10_-_2971407 | 0.21 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr23_-_20051369 | 0.21 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr4_+_14727018 | 0.21 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr16_-_13623928 | 0.21 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr24_+_1023839 | 0.20 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr14_+_23717165 | 0.19 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr10_+_29199172 | 0.19 |
ENSDART00000148828
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr25_+_10410620 | 0.19 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr2_-_32826108 | 0.19 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr25_-_32869794 | 0.19 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr21_-_26490186 | 0.18 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr19_-_32914227 | 0.18 |
ENSDART00000186115
ENSDART00000124246 |
mtdha
|
metadherin a |
| chr23_-_7799184 | 0.18 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr4_+_14727212 | 0.18 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr8_-_31107537 | 0.18 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr16_-_29194517 | 0.17 |
ENSDART00000046114
ENSDART00000148899 |
mef2d
|
myocyte enhancer factor 2d |
| chr18_+_1837668 | 0.17 |
ENSDART00000164210
|
CABZ01079192.1
|
|
| chr13_+_31205439 | 0.15 |
ENSDART00000132326
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr12_+_22580579 | 0.15 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr5_+_63857055 | 0.13 |
ENSDART00000138950
|
rgs3b
|
regulator of G protein signaling 3b |
| chr11_-_29768054 | 0.12 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr5_+_27525477 | 0.12 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr11_+_18873113 | 0.11 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr23_+_30835589 | 0.10 |
ENSDART00000154145
|
npbwr2a
|
neuropeptides B/W receptor 2a |
| chr7_-_12464412 | 0.09 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr25_+_6471401 | 0.09 |
ENSDART00000132927
|
snx33
|
sorting nexin 33 |
| chr12_+_25432627 | 0.09 |
ENSDART00000011662
|
ppm1bb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Bb |
| chr19_-_38830582 | 0.08 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr15_-_755023 | 0.08 |
ENSDART00000155594
|
znf1011
|
zinc finger protein 1011 |
| chr4_+_3980247 | 0.08 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr9_-_35633827 | 0.07 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr16_-_22930925 | 0.07 |
ENSDART00000133819
|
si:dkey-246i14.3
|
si:dkey-246i14.3 |
| chr2_-_26720854 | 0.07 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr9_-_3934963 | 0.06 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr20_+_13783040 | 0.06 |
ENSDART00000115329
ENSDART00000152497 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr22_-_24284447 | 0.06 |
ENSDART00000149894
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr14_-_7207961 | 0.06 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr19_-_32641725 | 0.06 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr24_+_36452927 | 0.05 |
ENSDART00000168181
|
zgc:114120
|
zgc:114120 |
| chr18_-_37007294 | 0.05 |
ENSDART00000088309
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr18_-_19095141 | 0.05 |
ENSDART00000090962
ENSDART00000090922 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr22_+_16308450 | 0.05 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr20_+_23185530 | 0.05 |
ENSDART00000142346
|
lrrc66
|
leucine rich repeat containing 66 |
| chr16_-_37415797 | 0.04 |
ENSDART00000188823
|
si:ch211-208k15.1
|
si:ch211-208k15.1 |
| chr12_+_20412564 | 0.04 |
ENSDART00000186783
|
arhgap17a
|
Rho GTPase activating protein 17a |
| chr9_+_1313418 | 0.04 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr16_-_30903930 | 0.04 |
ENSDART00000143996
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr7_+_30787903 | 0.03 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr23_+_26946744 | 0.03 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr18_-_37007061 | 0.03 |
ENSDART00000136432
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr21_-_43949208 | 0.03 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr2_-_54946834 | 0.02 |
ENSDART00000192280
|
CABZ01086574.1
|
|
| chr11_-_19775182 | 0.02 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr11_-_39105253 | 0.02 |
ENSDART00000102827
|
p3h1
|
prolyl 3-hydroxylase 1 |
| chr21_+_11244068 | 0.02 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr21_+_15824182 | 0.02 |
ENSDART00000065779
|
gnrh2
|
gonadotropin-releasing hormone 2 |
| chr9_-_32730487 | 0.02 |
ENSDART00000147795
|
olig1
|
oligodendrocyte transcription factor 1 |
| chr4_-_11106856 | 0.02 |
ENSDART00000150764
|
BX784029.3
|
|
| chr24_-_29963858 | 0.02 |
ENSDART00000183442
|
CR352310.1
|
|
| chr4_-_75681004 | 0.02 |
ENSDART00000186296
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr15_-_37835638 | 0.02 |
ENSDART00000128922
|
si:dkey-238d18.3
|
si:dkey-238d18.3 |
| chr7_-_54679595 | 0.02 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr22_+_16308806 | 0.02 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr5_-_51619742 | 0.01 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr17_-_29192987 | 0.01 |
ENSDART00000164302
|
sptbn5
|
spectrin, beta, non-erythrocytic 5 |
| chr22_-_7129631 | 0.01 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr9_+_11034314 | 0.01 |
ENSDART00000032695
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr9_-_44897553 | 0.01 |
ENSDART00000110697
|
dnajc10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr20_-_27225876 | 0.00 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr7_-_17591007 | 0.00 |
ENSDART00000171023
|
CU672228.1
|
|
| chr11_+_13224281 | 0.00 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
Network of associatons between targets according to the STRING database.
First level regulatory network of dlx3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 0.8 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.3 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.4 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.8 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.2 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.3 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 1.2 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.4 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.3 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.6 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.4 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 3.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.6 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |